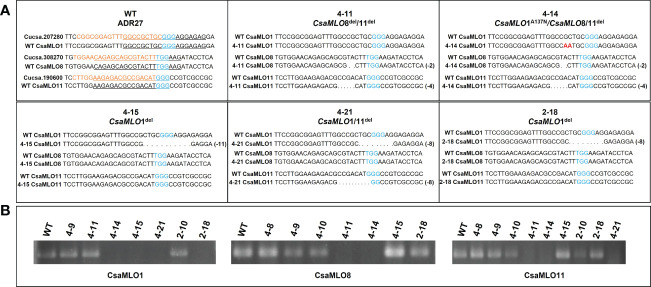
Figure 2.
Nucleotide sequence alignment of CsaMLOs. Clade V CsaMLOs of WT-ADR27 were aligned with the reference sequence to detect any genotypic differentiation, and the underlined nucleotides on the alignment shows the PCR primers for detection of homozygous mutants; (A) the sequence results indicated that 4-11 has deletion mutations on CsaMLO8 (2 bp) and CsaMLO11 (4 bp) even if there is no mutation on the CsaMLO1. Although 4-14 is triple mutant (2 bp del at CsaMLO8 and 4 bp del at CsaMLO11), there is substitution mutation on CsaMLO1 with 2 nucleotides change as “AA” rather than “GC” on the Cas9 cleavage site. 4-15 and 2-18 were determined as CsaMLO1 del with 11 bp and 8 bp deletions as a result of NHEJ. Also 8 bp deletions were detected on 4-21’s CsaMLO1 and CsaMLO11; (B) The PCR products were observed to be non-mutant genotype in terms of CsaMLO. However, there was no amplification in PCR results of mutant, because of homozygous deletion or substitution on the primer binding sites.