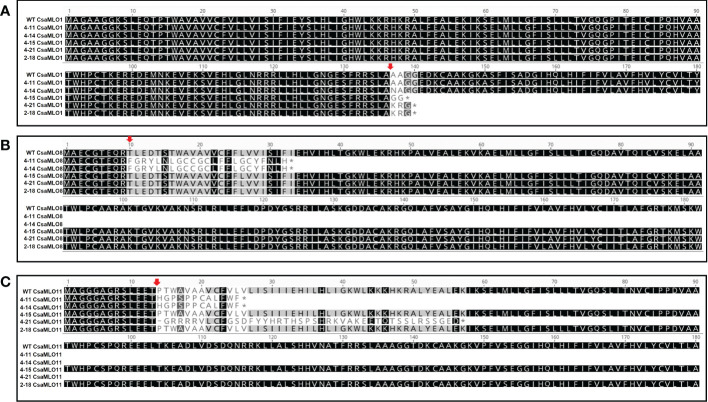
Figure 3.
Amino acid alignment using sequences of mutants and WT plants. (A) When the deletions on the Cas9 cleavage (red arrow) cause stop codon formation on the 4-15’s CsaMLO1 139th amino acid; the stop codon formation was detected on 2-18 and 4-21 CsaMLO1’s 140th amino acid (3rd exon). The Alanine (A) was altered with Asparagine (N) at the 137th amino acid as a result of the NHEJ caused by Cas9 cleavage at the gRNA1 target. (B) 2 bp deletions cause the unexpected stop codon formation on the 33rd amino acid of CsaMLO8 in 4-11 and 4-14. The target site of gRNA2 was on the first exon for CsaMLO8 and the changed amino acid structures are at the Cas9 cleavage (10th aa) site for this gene. The early stop codon formation caused the loss-of CsaMLO8 function in 4-11 and 4-14. (C) The mutations seem to be responsible for the stop codon formation in CsaMLO11 and stop codon’s positions of 4-11 and 4-14’s CsaMLO11 closer to that gRNA3 than 4-15 and 2-18 CsaMLO11. However, ORF shifts are initiated at gRNA3 position in whole CsaMLO11 del.