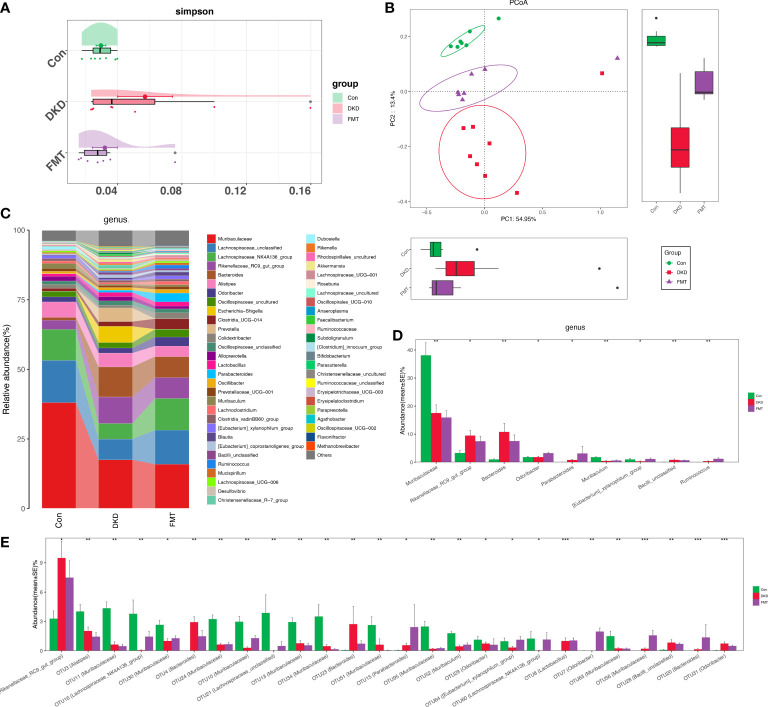
Figure 7.
Bacterial diversity and compositional alteration analysis of mouse models. Simpson indices (A) and PCoA analysis (B) were used to assess α and β diversity, respectively. PCoA analysis was measured by unweighted UniFrac distance at OTU level. Adonis revealed that unweighted analysis taking OTU abundance into account could better reflect the spatial differences among three groups (R 2 = 0.2233, p = 0.0001). Intestinal microbial composition at the genus level (C). Kruskal–Wallis rank-sum compares and identifies significantly altered bacteria at the genus and OTU level and the genus (D) or OTU (E) with gradually increased or decreased abundance between three groups was shown.