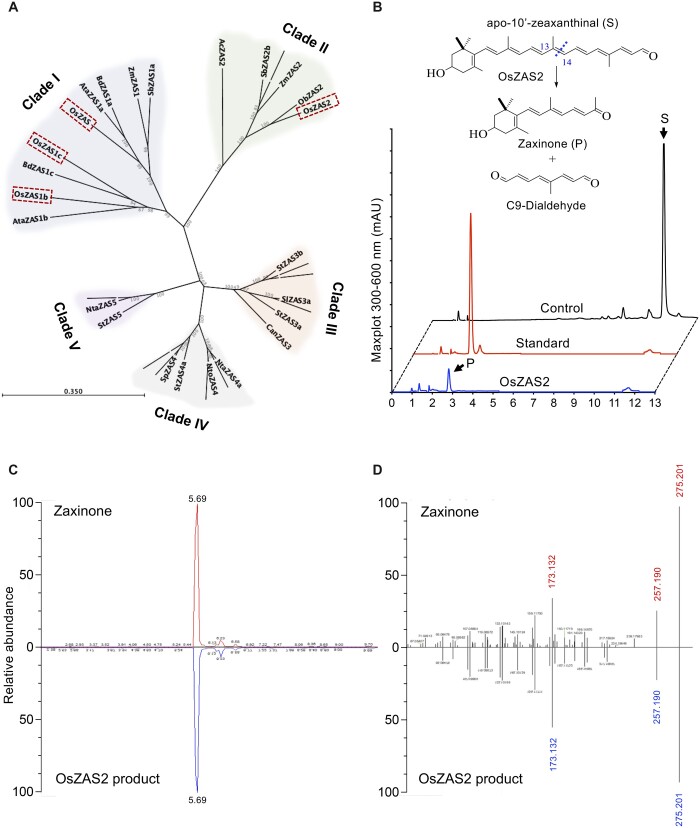
Figure 1.
Phylogenetic analysis of ZAS enzymes and analysis of OsZAS2 enzymatic activity. A, Phylogenetic tree analysis of ZAS orthologs from selected monocot and dicot plants, showing bootstrap values on nodes of each cluster. Dashed rectangles represent rice ZAS members. The scale bar represents the number of amino acid change per site. B, HPLC chromatogram of in vitro incubation of OsZAS2 with apo-10′-zeaxanthinal (I) yielded zaxinone (II) and a presumed C9-dialdehyde. The maximum absorbance (mAU) peak for substrate and product is shown at 347 and 450 nm (mAU), respectively. The representation of zaxinone production in the Figure B adapted from Wang et al. (2019), which is permitted to adaptation under a Creative Commons Attribution 4.0 International License. C, Verification of OsZAS2 in vitro product, based on retention time, (D) accurate mass and MS/MS pattern and in comparison to zaxinone standard.