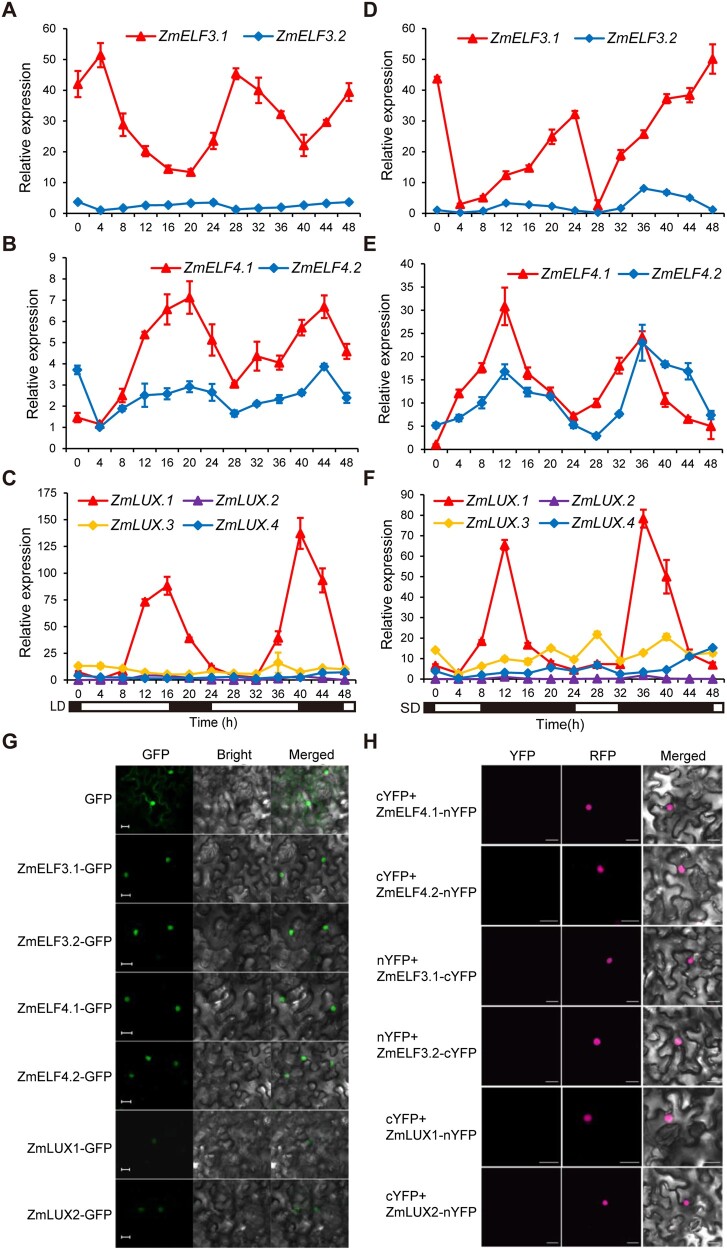
Figure 2.
Expression pattern of ZmELF3s, ZmELF4s, and ZmLUXs and subcellular localization of their encoded proteins. A–F, ZmELF3.1/3.2, ZmELF4.1/4.2, and ZmLUX1/2/3/4 all show a rhythmic diurnal expression pattern under both LD (A–C) and SD (D–F) conditions. The sixth leaves from V6-stage plants of ZC01 (a temperate line) grown under the indicated conditions were used for RNA extraction and RT-qPCR. Values are means ± SD (n = 3 technical replicates). Two independent experiments were performed and the results were similar (leaves from six different plants were used for each biological replicate). The black bars and white bars indicate the dark period and the light period, respectively. G, ZmELF3.1/3.2, ZmELF4.1/4.2, and ZmLUX1/2 proteins localize to the nucleus. The expression constructs GFP, ZmELF3.1/3.2-GFP, ZmELF4.1/4.2-GFP, and ZmLUX1/2-GFP were individually infiltrated into N. benthamiana leaves and then incubated for 48 h in the dark prior to imaging using a confocal microscope. Scale bars, 20 µm. Three independent experiments were performed, with similar results. H, Negative controls for the BiFC assay for the interaction between ZmELF3.1/3.2, ZmELF4.1/4.2, and ZmLUX1/2 proteins. The nuclear marker AHL-RFP indicates the nuclei in N. benthamiana leaf epidermal cells. Scale bars, 20 µm. Three independent experiments were performed, with similar results.