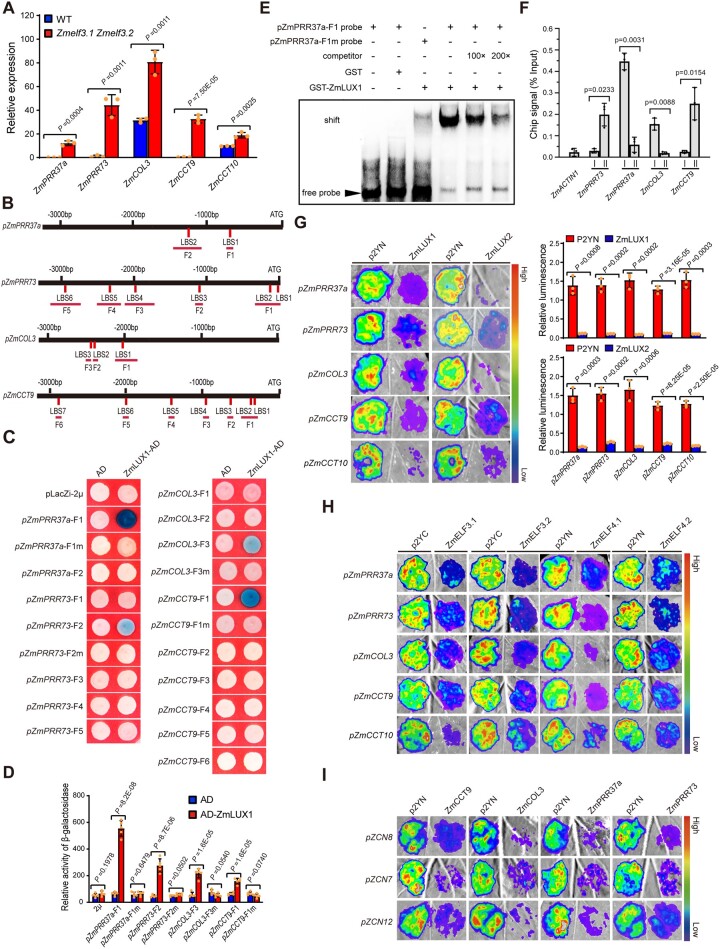
Figure 4.
Maize EC directly represses the expression of several flowering repressor genes. A, RT-qPCR assay showing that the expression levels of several flowering repressor genes, ZmPRR37a, ZmPRR73, ZmCOL3, ZmCCT9, and ZmCCT10, are significantly upregulated in the Zmelf3.1 Zmelf3.2 double mutant compared to the WT (ZC01). Values are means ± SD (n = 3 technical replicates). Two biological replicates were performed and the results were similar (leaves from six plants were used for each biological replicate). B, The promoters of the above flowering repressor genes harbor multiple typical LUX binding sites (LBS, 5′-GATWCG-3′, 5′-GATWKG-3′, or 5′-GATWCY-3′, where W indicates A or T, K indicates C or T, and Y indicates G or T). The positions of the LBS motifs in each promoter are indicated with red vertical lines and the promoter fragments used for Y1H assay are marked with red horizontal lines. C, Y1H assay showing that ZmLUX1 directly binds to specific promoter fragments containing the LBS motif of ZmPRR37a (5′-CGTATCGTATC-3′), ZmPRR73 (5′-GATTCG-3′), ZmCOL3 (5′-GATTCG-3′), and ZmCCT9 (5′-AGAATC CATATC-3′). For mutagenesis, the LBS motif was mutated to 5′-TCCAAGTGATG-3′ in ZmPRR37apro-F1, 5′-CACACA-3′ in ZmPRR73pro-F2, 5′-CTAGGA-3′ in ZmCOL3pro-F3, and 5′-CTAGGA TCCTAG-3′ in ZmCCT9pro-F1. Three independent experiments showed similar results. D, Quantification of ZmLUX1 binding activity. Five independent yeast clones were used for activity determination. Values are means ± SD (n = 5 independent clones). Significant differences were determined by the Student’s t test. E, EMSA showing that recombinant GST-ZmLUX1 fusion protein binds to a biotin-labeled probe of the ZmPRR37a promoter. GST protein was used as a negative control. Two independent experiments were performed, with similar results. F, ChIP-qPCR assay of enrichment for ZmELF3.1 at the ZmPRR37a, ZmPRR73, ZmCOL3, and ZmCCT9 promoters in the leaves of V2-stage Ubipro:ZmELF3.1-EGFP transgenic seedlings grown under LD conditions. The “I” and “II” represent the individual amplified fragment of each promoter as shown in (B). Values are means ± SD (n = 3 biological replicates, leaves from six plants were used for each replicate). P-value was determined by the Student’s t test. Two independent experiments showed similar results. G, Transient expression assay showing that ZmLUX1 and ZmLUX2 repress the expression of the above flowering repressor genes. Values are means ± SD (n = 3 different infiltrated regions). At least six independent experiments were performed, with similar results. Representative images of N. benthamiana leaves 72 h after infiltration are shown. Significant differences were determined by the Student’s t test. H, Transient expression assay shows that both ZmELF3s and ZmELF4s suppress the expression of ZmPRR37a, ZmPRR73, ZmCOL3, and ZmCCT9. At least six independent experiments were performed, with similar results. Representative images of N. benthamiana leaves 72 h after infiltration are shown. I, Transient expression assay shows that ZmPRR37a, ZmPRR73, ZmCOL3, and ZmCCT9 suppress the expression of ZCN8, ZCN7, and ZCN12. At least six independent experiments were performed, with similar results. Representative images of N. benthamiana leaves 72 h after infiltration are shown.