Abstract
Background/Aim: Brain metastases (BMs) are the most frequent intracranial tumors in adults and one of the greatest challenges for modern oncology. Most are derived from lung, breast, renal cell, and colorectal carcinomas and melanomas. Up to 14% of patients are diagnosed with BMs of unknown primary, which are commonly characterized by an early and aggressive metastatic spread. It is important to discover novel biomarkers for early identification of BM origin, allowing better management of patients with this disease. Our study focused on microRNAs (miRNAs), which are very stable in frozen native and FFPE tissues and have been shown to be sensitive and specific diagnostic biomarkers of cancer. We aimed to identify miRNAs with significantly different expression in the five most frequent groups of BMs and develop a diagnostic classifier capable of sensitive and specific classification of BMs.
Materials and Methods: Total RNA enriched for miRNAs was isolated using the mirVana miRNA Isolation Kit from 71 fresh-frozen histopathologically confirmed BM tissues originating in 5 cancer types. Sequencing libraries were prepared using the QIAseq miRNA Library Kit and sequenced on the NextSeq 500 platform. MiRNA expression was further validated by RT-qPCR.
Results: Differential analysis identified 373 miRNAs with significantly different expression between 5 BM groups (p<0.001). A classifier model was developed based on the expression of 6 miRNAs (hsa-miR-141-3p, hsa-miR-141-5p, hsa-miR-146a-5p, hsa-miR-194-5p, hsa-miR-200b-3p and hsa-miR-365b-5p) with the ability to correctly classify 91.5% of samples. Subsequent validation confirmed both significantly different expression of selected miRNAs in 5 BM groups as well as their diagnostic potential.
Conclusion: To date, our study is the first to analyze miRNA expression in various types of BMs using small RNA sequencing to develop a diagnostic classifier and, thus, to help stratify BMs of unknown primary. The presented results confirm the importance of studying the dysregulated expression of miRNAs in BMs and the diagnostic potential of the validated 6-miRNA signature.
Keywords: Brain metastases, microRNA, small RNA sequencing, classifier, diagnosis
Metastatic tumors present one of the most challenging issues in modern oncology as they are very detrimental to patients’ quality of life. Among the most destructive metastatic lesions are brain metastases (BM), which are the most frequent intracranial tumors in adults and their management represents one of the most urgent unmet clinical needs in the care for cancer patients. The higher incidence of BMs may be caused by the advances in tumor therapy and diagnostics as well as supportive care, since the increasingly more efficient therapy prolongs the overall survival of patients but simultaneously raises the probability of metastatic spread while the higher sensitivity and availability of imaging techniques allows detection of small asymptomatic metastases, which would previously go unnoticed (1). Despite these advancements, BMs remain a fatal event in cancer progression and often lead to an overall survival of 16 months or less since the diagnosis of metastatic growth. Generally, the median overall survival of patients with BMs highly varies from 3-17 months for patients with BMs derived from gastrointestinal cancer to 7-46 months for patients with BMs originating in lung carcinomas (2). BMs were previously found by various autopsy studies to be partly or entirely the cause of death in approximately 25% of all cases (1), although these studies tend to be outdated and the autopsy rates have been overall on the decline during the past three decades (3). According to more recent literature, the estimates of BM incidence vary approximately between 0.07% and 19.9%, depending on the primary cancer site and subtype (1,4). Out of these cases, most are derived from tumors with a high tendency to metastasize to brain, including lung carcinomas (20-56%), breast carcinomas (5-20%), melanomas (7-16%), renal cell carcinomas (2-9%), and colorectal carcinomas (2-9%), while BMs derived from tumors of esophagus, liver, pancreas, stomach, endometrium, ovaries, prostate, testes, and bladder are less common (1,5-9). Between 1 to 14% of patients are diagnosed with BMs of unknown primary (BMUP), when the primary cancer diagnosis is unknown, and the identification of primary tumor is impossible (6,7). BMUP are commonly characterized by an early and aggressive metastatic spread, and they respond poorly to conventional therapy, resulting in a dismal prognosis (10,11).
Since early identification of the original primary site in patients with BMUP would certainly be beneficial for administering more effective therapy, it is important to search for novel biomarkers for a precise stratification of patients with BMs. MicroRNAs (miRNAs), a well-known subclass of small non-coding RNAs, which function as posttranscriptional regulators of gene expression, bear certain advantages over other biomarkers. Firstly, the high dysregulation of their expression has been frequently observed and extensively studied in various types of malignancies, promoting the search for a miRNA-based signature for many diseases, including BMs. Secondly, the technology for detection of miRNAs using small RNA sequencing and real-time PCR coupled with reverse transcription (RT-qPCR) in many types of biological specimen has been well-established by now, facilitating a precise quantification of miRNA expression. Unlike mRNAs, which need to be further translated into proteins to exert their function, mature miRNAs are already fully functional molecules, and it is therefore reasonable to consider the expression quantified by RT-qPCR to be very close to the real expression in tissues. Thirdly, due to their short length and structure, most miRNAs are generally very stable and resistant to changes in temperature and pH; this allows for the detection of miRNAs in both frozen native specimen and archived biological material, such as formalin-fixed and paraffin-embedded (FFPE) samples (12). Lastly, they have previously been shown to possess the potential to be diagnostic, prognostic, and predictive biomarkers, as well as therapeutic targets of BMs (13). Therefore, a classifier based on specific patterns of miRNA expression in BMs of various origins could serve as a promising diagnostic tool for determining both the original primary site and the prognosis of patients with BMUP. In our present study, we aimed to analyze miRNA expression in BMs with origin in lung carcinomas (BML), breast carcinomas (BMB), melanomas (BMM), renal cell carcinomas (BMR), and colorectal carcinomas (BMC) using small RNA sequencing to identify miRNAs with significantly different expression and develop miRNA-based diagnostic classifier capable of sensitive and specific classification of BMs with origin in these five primary cancer types. The gathered data could then lay the groundwork for a future robust classifier capable of precise stratification of BMs with various origin, including BMUP.
Materials and Methods
Patient samples. Native BM tissue samples were collected by cooperating neurosurgical departments of University Hospital Brno and St. Anne’s University Hospital Brno (Brno, Czech Republic) during surgery, which is a part of the standard treatment protocol. The study and the informed consent form were approved by the research ethics committee of University Hospital Brno under the code EKFNB-17-06-28-01. A signed informed consent form was obtained from each patient prior to the beginning of the treatment and the collection of patient tissue samples. The study methodologies conformed to the standards set by the Declaration of Helsinki. Seventy-one fresh tissue samples collected for the exploratory phase of the study were immediately stored after the collection in RNAlater Stabilization Solution (Thermo Fisher Scientific, Waltham, MA, USA) at 4˚C for 24 h and then frozen at –80˚C until further use. The retrospective cohort used in the validation phase of the study consisted of 119 FFPE samples, which were supplied by the cooperating pathology departments of University Hospital Brno, St. Anne’s University Hospital Brno (Brno, Czech Republic), and University Hospital Hradec Králové (Hradec Králové, Czech Republic). The histology of all fresh and FFPE tissue samples collected for this study was analyzed according to the WHO 2021 classification scheme independently by two histopathologists who confirmed that these samples belonged to the 5 most frequent types of BMs.
RNA isolation and purification. Fresh-frozen tissue samples were mechanically homogenized with 1.4 mm ceramic beads (Qiagen, Hilden, Germany) in the presence of Lysis/Binding Buffer from mirVana miRNA Isolation Kit (Thermo Fisher Scientific) in the Precellys Evolution homogenizer (Bertin Instruments, Montigny-le-Bretonneux, France). Total RNA enriched for small RNA species was then isolated and purified using the mirVana miRNA Isolation Kit according to the manufacturer’s instructions. Serial sections were cut from each paraffin-embedded tissue block using a microtome (eight 10 μm thick slices per block) and stored in 1.5 ml tubes until further use. The sections were then deparaffinized by 1 ml of xylene per tube and mixed on a vortex mixer. Samples were centrifuged at the maximum speed for 2 min, the supernatant was discarded, and the xylene treatment was repeated once. Then 1 ml of absolute ethanol was added to the tissue pellet, the sample was mixed on a vortex mixer and centrifuged at the maximum speed for 2 min. The supernatant was discarded, and the ethanol treatment was repeated once. The ethanol residue was left to evaporate at 37˚C and the tissue pellet was digested using the mixture of 180 μl of ATL buffer and 20 μl of proteinase K (both Qiagen) at 55˚C overnight. The sample was mixed with 600 μl of Lysis/Binding Buffer and 100 μl of miRNA Homogenate Additive, incubated for 10 min on ice, and then mixed with 600 μl of Acid Phenol:Chloroform:IAA (125:24:1), pH 4.5 (all Thermo Fisher Scientific) and centrifuged at maximum speed for 5 min. Total RNA enriched for small RNA species was then isolated and purified using the mirVana miRNA Isolation Kit according to the manufacturer’s instructions.
Nucleic acid quantity and quality control. Purified RNA was quantified using the NanoDrop 2000 spectrophotometer, and the absorbance ratios A260/A280 and A260/A230, indicating RNA purity, were automatically calculated using the NanoDrop 2000 software (version 1.6.0.198). Concentrated RNA samples were subsequently diluted for downstream analyses and quantified precisely in Qubit 2.0 Fluorometer using Qubit RNA BR Assay Kit (both Thermo Fisher Scientific). The RNA integrity was assessed by capillary gel electrophoresis using Agilent 2200 TapeStation and Agilent RNA ScreenTape System (both Agilent Technologies, Santa Clara, CA, USA) and quantified as the RNA integrity number (RIN) by the 2200 TapeStation analysis software (version 2.1.27.8350).
Library preparation, pooling, and sequencing. Seventy-one total RNA samples with RIN ≥5.7 (median 8.2) were used as a template for the construction of small RNA libraries using QIAseq miRNA Library Kit (Qiagen). All procedures were performed according to the manufacturer’s protocol. The input RNA amount was 100 ng. The concentration of prepared libraries was measured using Qubit 2.0 Fluorometer and Qubit dsDNA HS Assay Kit (both Thermo Fisher Scientific). Libraries were also analyzed using Agilent 2200 TapeStation and Agilent High Sensitivity D1000 ScreenTape System (both Agilent Technologies) and then pooled in equimolar ratio based on their molarity, which was calculated using online weight-to-moles conversion calculator for nucleic acids. Library pools (23 to 24 libraries in each pool) were then processed according to the NextSeq System Denature and Dilute Libraries Guide (14). Denatured and diluted PhiX Control v3 was added at 1% to all pools as an internal standard and single-read sequencing with 75 bp read length was performed using NextSeq 500 Sequencing System and NextSeq 500/550 High Output v2 kit (75 cycles) (all Illumina, San Diego, CA, USA).
Processing of small RNA sequencing data. The pre-alignment quality control (QC) of the sequencing data was done using FastQC (version 0.11.9) (15). Adaptors present within sequenced reads were trimmed off with cutadapt (version 3.3) (16). Adapter-trimmed small RNA sequencing reads were collapsed exploiting unique molecular identifiers (UMIs) with FASTX-Toolkit (version 0.0.14) (17). Subsequently, reads were quality trimmed using cutadapt and reads shorter than 15 bp were removed from the dataset. The remaining reads were mapped against the database miRBase (version 21) (18) using the miraligner tool (version 3.2) (19). All generated numerical and graphical output from QC was gathered in cohesive reports via MultiQC (version 1.7) (20). All statistical analyses were performed in the R environment (version R4.0.3).
Differential expression analysis. Differential expression analysis was carried out using the Bioconductor (version 3.11) package limma (version 3.44.3) (21). The complete linkage (farthest neighbor clustering) method with euclidean distance measure was used for the unsupervised clustering. The expression levels of miRNA with more than 1 count per million in at least 5 samples were analyzed and compared between 5 groups of BMs. Results were summarized in heatmaps with clustergram. MiRNAs with significantly different expression were identified by having Benjamini-Hochberg adjusted p-Value smaller than 0.05.
Development of a classifier based on miRNA expression. The discovery of miRNA biomarker signature was performed using the Boruta algorithm (version 7.0.0) (22), followed by multinomial regression models (unordered categories of original tumors). The selection of a subset of miRNAs with predictive ability was done by leave-one-out cross-validation. The top 100 miRNAs from limma F-test were selected as candidate predictors. For each patient, the Boruta algorithm was employed on a testing set of 70 remaining patients which led to a subset of important miRNAs. The predictive ability of the selected model was validated by the excluded patient. If the prediction failed, i.e., the multinomial regression model incorrectly predicted the tumor origin of the excluded patient, the whole model was discarded. The same procedure was repeated 71 times and a consensus model was formed from all validated miRNA signatures. For practical reasons, the consensus model was then collapsed to a minimal signature without compromising its predictive ability.
Validation of sequencing results. Total RNA isolated from FFPE sections was reversely transcribed to cDNA using TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The expression levels were then analyzed by quantitative PCR (RT-qPCR) using TaqMan Universal Master Mix II and TaqMan MicroRNA Assays for hsa-miR-16-5p (000391), hsa-miR-141-3p (000463), hsa-miR-141-5p (002145), hsa-miR-146a-5p (000468), hsa-miR-194-5p (000493), hsa-miR-200b-3p (002251) and hsa-miR-365b-5p (121213_mat) (all Thermo Fisher Scientific) on a LightCycler 480 Instrument II (Roche, Basel, Switzerland). All PCR reactions were performed in technical duplicates. Expression levels were normalized for expression of hsa-miR-16-5p, which was chosen as a normalizer based on its stable and high expression across all samples from the exploratory phase (Benjamini-Hochberg adjusted p-Value of 0.57; average normalized expression of 16.18). Relative expression levels were compared between each pair of groups or all groups in Prism 8 software (GraphPad Software, San Diego, CA, USA) using the Mann-Whitney U-test or Kruskal-Wallis one-way ANOVA, respectively. Finally, the predictive ability of the previously established model based on the expression profiles of six miRNAs was validated on the qPCR data as an independent dataset.
Results
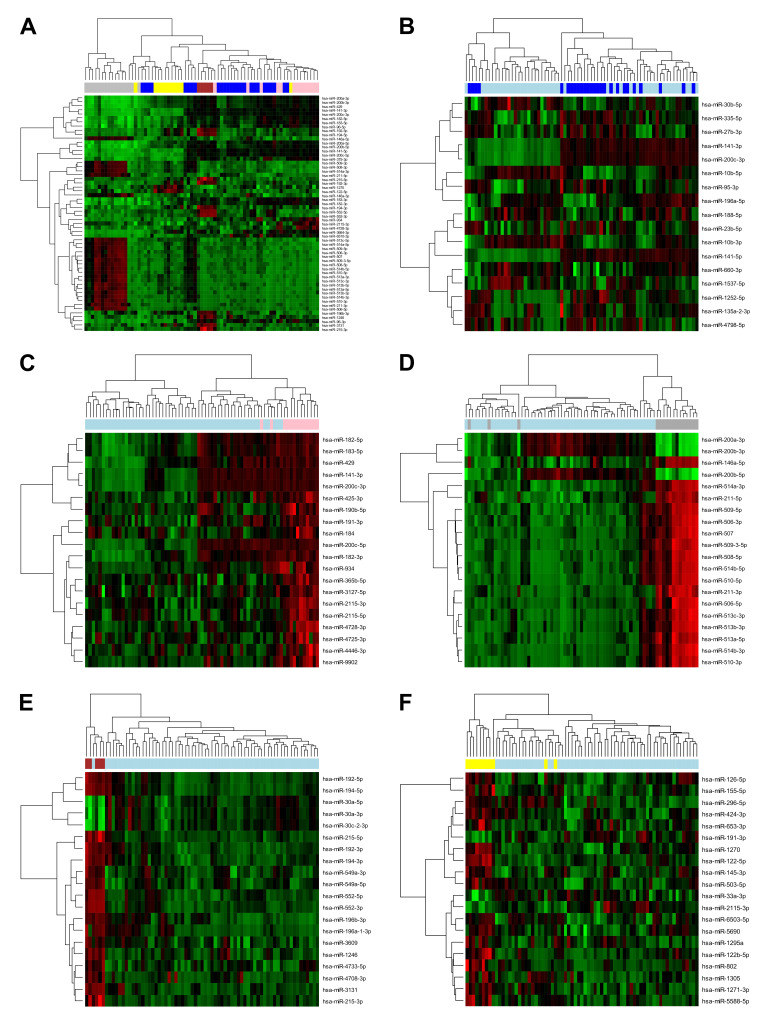
Sequencing analysis of microRNA expression in five most prevalent groups of brain metastases identifies differentially expressed microRNAs. In the exploratory phase of our study, we performed a small RNA sequencing analysis to identify miRNAs with significantly different expression profiles in 71 BM samples belonging to 5 groups based on the histology of the primary tumors from which the metastases originated, namely BML (37%), BMM (23%), BMB (18%), BMR (15%), and BMC (7%). Out of the 1,091 miRNAs, which were taken into the analysis, 373 miRNAs were significantly differentially expressed among the 5 groups (Benjamini-Hochberg adjusted p-Value <0.05), with 58 miRNAs having adjusted p-Values smaller than 0.000001 (Figure 1A). Moreover, each BM group was also compared with another group consisting of the other BMs to identify sets of miRNAs with unique expression pattern in the individual groups of BMs (Figure 1B-F).
Figure 1. Results from differential analysis of miRNA expression assessed by small RNA sequencing in 71 samples of brain metastases (BM) derived from 5 tumor types, including lung carcinomas (dark blue), breast carcinomas (pink), melanomas (gray), colorectal carcinomas (dark red), and renal cell carcinomas (yellow). (A) The miRNA expression profiles in the 5 groups of BMs were compared, resulting in the identification of 58 significantly differentially expressed miRNAs with Benjamini-Hochberg adjusted p-Value smaller than 0.000001. The expression profiles of these miRNAs were visualized in a heatmap with dendrogram. Furthermore, each group was also compared with another group comprised of other BMs. The heatmaps with dendrogram show the results of hierarchical clustering based on the expression of (B) 17 differentially expressed miRNAs (adjusted p-Value <0.05) in BMs derived from lung carcinomas (dark blue) vs. other BMs (light blue), and the 20 most significantly differentially expressed miRNAs in (C) BMs derived from breast carcinomas (pink) vs. other BMs (light blue) (adjusted p-Value ≤0.0017), (D) melanoma-derived BMs (gray) vs. other BMs (light blue) (adjusted p-Value <0.0001), (E) BMs derived from colorectal carcinomas (dark red) vs. other BMs (light blue) (adjusted p-Value ≤0.0002), and (F) BMs derived from renal cell carcinomas (yellow) vs. other BMs (light blue) (adjusted p-Value ≤0.0135). The green-black-red spectrum represents miRNA expression in samples from low (green) to high expression (red).
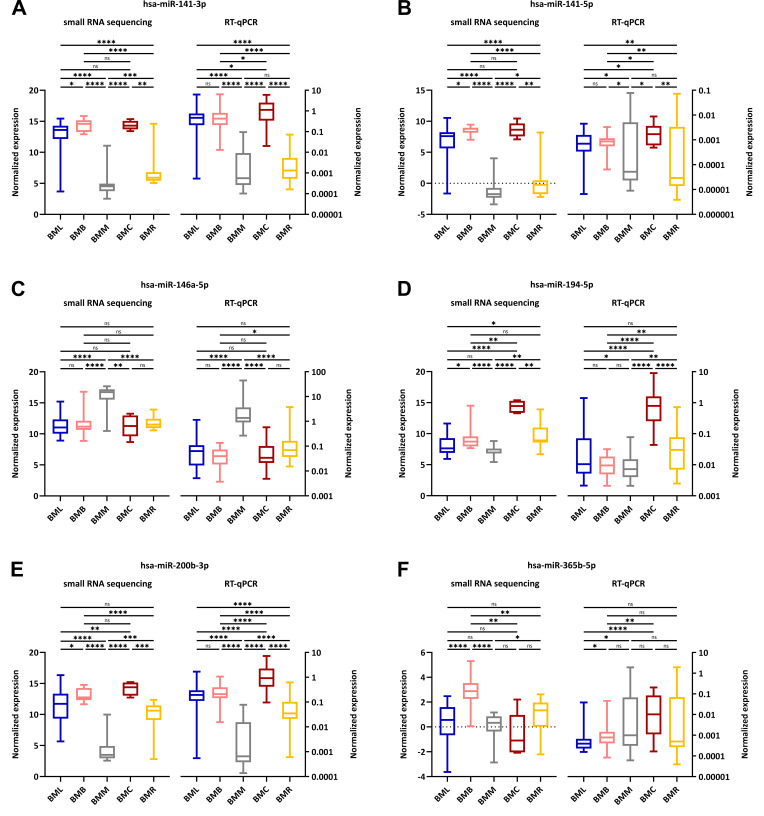
Six-microRNA signature comprises a classifier with a diagnostic potential for the five major groups of brain metastases. To establish a model with the ability of predicting the origin of BMs, the top 100 differentially expressed miRNAs in the 5 groups of BMs (adjusted p-Value <0.0005) were taken into the analysis using the Boruta algorithm followed by multinomial regression models. Using this approach, 43 miRNA signatures were obtained and validated. Finally, a consensus model was formed from 32 miRNAs occurring in each of the obtained signatures, consisting of hsa-miR-10b-5p, hsa-miR-122-5p, hsa-miR-141-3p, hsa-miR-141-5p, hsa-miR-146a-5p, hsa-miR-155-5p, hsa-miR-183-3p, hsa-miR-188-5p, hsa-miR-191-3p, hsa-miR-194-5p, hsa-miR-200a-3p, hsa-miR-200a-5p, hsa-miR-200b-3p, hsa-miR-200b-5p, hsa-miR-200c-3p, hsa-miR-200c-5p, hsa-miR-211-3p, hsa-miR-211-5p, hsa-miR-215-5p, hsa-miR-30a-5p, hsa-miR-365a-3p, hsa-miR-365b-5p, hsa-miR-375-3p, hsa-miR-429, hsa-miR-514a-3p, hsa-miR-514a-5p, hsa-miR-2115-3p, hsa-miR-2115-5p, hsa-miR-3131, hsa-miR-4728-3p, hsa-miR-4780 and hsa-miR-6510-3p. For validation, with practical and economical aspects in mind, the model was then collapsed to a minimal signature consisting of 7 miRNAs (hsa-miR-141-3p, hsa-miR-141-5p, hsa-miR-146a-5p, hsa-miR-194-5p, hsa-miR-200b-3p, hsa-miR-365b-5p, hsa-miR-4780). The predictive ability of this simplified model was confirmed by 100% accuracy, i.e., all samples were correctly classified. Hsa-miR-4780 was then excluded from the model because according to the miRBase, the confidence of its annotation and existence was very low, resulting in the conclusion that the probability of successful validation of its expression was also low. The final 6-miRNA classifier model had the ability to accurately classify 65 of 71 samples, i.e., 91.5% of all samples. Group-wise, the classifier was able to correctly classify 88.46% of BML, 84.62% of BMB, 100% of BMM, 100% of BMC, and 90.91% of BMR samples. The results from the classifier, the calculated values of sensitivity, specificity and 95% confidence interval, and the hierarchical clustering heatmap based on the expression of 6 miRNAs are shown in Figure 2.
Figure 2. Results from the classifier algorithm and the differential analysis based on the miRNA expression assessed by small RNA sequencing. (A) The classifier algorithm is based on the expression of 6 miRNAs (hsa-miR-141-3p, hsa-miR-141-5p, hsa-miR-146a-5p, hsa-miR-194-5p, hsa-miR-200b-3p, hsa-miR-365b-5p) in 71 samples of BMs derived from lung carcinomas (BML), breast carcinomas (BMB), melanomas (BMM), colorectal carcinomas (BMC), and renal cell carcinomas (BMR). The real BM origin is represented by columns and the classifier prediction by rows. The classifier correctly identified the origin of 91.5% of samples which are highlighted in green. Incorrectly classified samples are highlighted in yellow. The corresponding sensitivity, specificity and 95% confidence interval (CI) values for each group of BMs are shown in the bottom part. (B) The hierarchical clustering analysis based on the expression of the 6 miRNAs in 71 samples of BMs derived from lung carcinomas (dark blue), breast carcinomas (pink), melanomas (gray), colorectal carcinomas (dark red), and renal cell carcinomas (yellow) was visualized in a heatmap with dendrogram. The green-black-red spectrum represents miRNA expression in samples from low (green) to high expression (red).
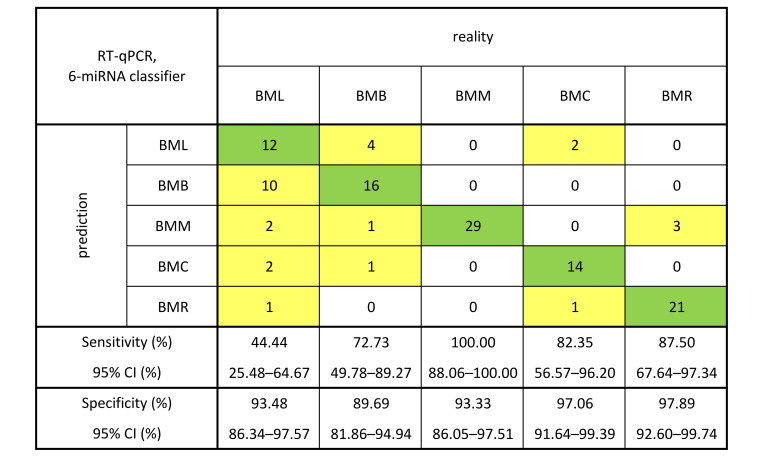
Validation in an independent cohort confirms the expression patterns of the six microRNAs and the diagnostic potential of the established model for all but one group of brain metastases. To validate the expression patterns obtained from the analysis of small RNA sequencing data and the established 6-miRNA signature and to rule out any effects of the specific sequencing platform, the expression levels of these miRNAs were measured using RT-qPCR in an independent cohort of 119 FFPE tissue samples of BMs [BML (22.7%), BMB (18.5%), BMM (24.4%), BMC (14.3%) and BMR (20.1%)]. The analysis of measured and normalized data using Mann-Whitney U-test confirmed the expression patterns observed in the exploratory phase of the study with notable deviation in the case of hsa-miR-365b-5p (Figure 3). Furthermore, Kruskal-Wallis one-way ANOVA also confirmed the significantly different expression of all 6 miRNAs in 5 groups of BMs (p<0.0001 for hsa-miR-141-3p, hsa-miR-146a-5p, hsa-miR-194-5p and hsa-miR-200b-3p; p=0.0007 for hsa-miR-141-5p; p=0.0035 for hsa-miR-365b-5p). The predictive ability of the previously established 6-miRNA signature was then tested on the RT-qPCR data as an independent dataset, and the model was shown to accurately classify most of the samples from all groups except BML, i.e., the classifier correctly classified 44% of BML, 73% of BMB, 100% of BMM, 82% of BMC, and 88% of BMR samples. The results from the classifier testing on RT-qPCR data and the calculated values of sensitivity, specificity and 95% confidence interval are shown in Figure 4.
Figure 3. Comparison of the expression patterns of the 6 miRNAs; namely (A) hsa-miR-141-3p, (B) hsa-miR-141-5p, (C) hsa-miR-146a-5p, (D) hsamiR-194-5p, (E) hsa-miR-200b-3p and (F) hsa-miR-365b-5p. The expression was analyzed by small RNA sequencing and RT-qPCR in 71 native tissue samples and 119 formalin-fixed paraffin-embedded tissue samples, respectively, which were derived from lung carcinomas (BML, blue), breast carcinomas (BMB, pink), melanomas (BMM, gray), colorectal carcinomas (BMC, dark red), and renal cell carcinomas (BMR, yellow). Median values of normalized expression were compared using the non-parametric Mann-Whitney U-test. The statistical significance of the results is denoted by ns (not significant; p>0.05), *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
Figure 4. Results from the classifier algorithm, which is based on the expression of 6 miRNAs (hsa-miR-141-3p, hsa-miR-141-5p, hsa-miR-146a-5p, hsa-miR-194-5p, hsa-miR-200b-3p, hsa-miR-365b-5p) assessed by RT-qPCR in 119 formalin-fixed paraffin-embedded tissue samples of brain metastases (BM). The real BM origin is represented by columns and the classifier prediction by rows. The classifier correctly identified the origin of 77.3% of samples which are highlighted in green. Incorrectly classified samples are highlighted in yellow. The corresponding sensitivity, specificity and 95% confidence interval (CI) values for each group of BMs are shown in the bottom part.
Discussion
BMs are a relatively frequent event in the later stages of primary solid cancers, leading to a generally dismal prognosis. The incidence of BMs has increased, possibly due to more efficient approaches to therapy, which on one hand prolong overall patient survival but on the other hand allow more time for metastasizing cells to invade and colonize the brain. Despite recent advances in the treatment and imaging of BMs, the prognosis of patients with BMs remains poor and is further exacerbated in the case of BMUP when the underlying origin of the disease is not known. It is, therefore, important to search for novel diagnostic molecular biomarkers and develop sensitive and specific classifier models capable of precise diagnostics of patients with BMs based on the expression of said biomarkers.
In our present study, we focused on miRNAs, which are very stable, well-researched and easily analyzable and they have been proven to be good diagnostic, prognostic, and predictive indicators for various diseases, including cancer (23). Using small RNA sequencing, we identified miRNAs with significantly different expression in the 5 major groups of BMs, including hsa-miR-200c-3p, hsa-miR-141-5p, hsa-miR-141-3p, hsa-miR-200c-5p, hsa-miR-215-5p, hsa-miR-200b-3p, hsa-miR-211-3p, hsa-miR-429, hsa-miR-200a-3p and hsa-miR-200b-5p, which were the ten most significantly differentially expressed miRNAs based on the corresponding adjusted p-Value. Out of these, eight (hsa-miR-141-3p, hsa-miR-141-5p, hsa-miR-200a-3p, hsa-miR-200b-3p, hsa-miR-200b-5p, hsa-miR-200c-3p, hsa-miR-200c-5p, hsa-miR-429) belong to the miR-200 family of miRNAs, which has been described to be involved in the inhibition of the epithelial-mesenchymal transition (EMT), the initiating step of cancer metastasis (24). More specifically, they enhance the expression of the cell-cell adhesion molecule E-cadherin by directly targeting and inhibiting its transcriptional repressors ZEB1 and ZEB2 (25,26). By contrast, they were found to promote colonization of secondary sites by metastasizing cells and formation of metastatic foci, the last steps of cancer metastasis, by inducing mesenchymal-epithelial transition (MET) (27). Interestingly, all 9 members of the miR-200 family (hsa-miR-141-3p, hsa-miR-141-5p, hsa-miR-200a-3p, hsa-miR-200a-5p, hsa-miR-200b-3p, hsa-miR-200b-5p, hsa-miR-200c-3p, hsa-miR-200c-5p, hsa-miR-429) were identified by the Boruta algorithm to be a part of the 32-miRNA signature with the ability to accurately classify all samples from 5 major groups of BMs. Furthermore, after identifying miRNAs with expression changes specific for each histological type of BM (Figure 1B-F), we found that, in total, 19 significantly differentially expressed miRNAs were also part of our 32-miRNA signature, namely hsa-miR-10b-5p, hsa-miR-141-3p, hsa-miR-141-5p, hsa-miR-188-5p, and hsa-miR-200c-3p, in the case of BML; hsa-miR-191-3p, hsa-miR-365b-5p, hsa-miR-2115-3p, hsa-miR-2115-5p and hsa-miR-4728-3p, in the case of BMB; hsa-miR-146a-5p, hsa-miR-200b-3p, hsa-miR-211-3p, and hsa-miR-211-5p, in the case of BMM; hsa-miR-194-5p, hsa-miR-215-5p, and hsa-miR-3131, in the case of BMC; and hsa-miR-122-5p, hsa-miR-155-5p, hsa-miR-191-3p and hsa-miR-2115-3p, in the case of BMR.
After collapsing the 32-miRNA signature, hsa-miR-141-3p, hsa-miR-141-5p and hsa-miR-200b-3p remained in the 6-miRNA model. The collapsing was done out of practicality since it would be financially unachievable to validate the expression of all 32 miRNAs using RT-qPCR in 119 samples. The 6-miRNA classifier was able to correctly classify 91.5% samples which we deemed to be sufficient. The model was also proven to be highly sensitive and specific since the calculated values of sensitivity and specificity for all 5 groups were higher than 84% and 93%, respectively. The expression of these 6 miRNAs was, therefore, proven to be a very good indicator of the primary origin of analyzed BM samples. We then proceeded to validate the results from the exploratory phase in an independent validation cohort and confirmed the overall expression trends observed in the exploratory phase of the study. The only notable exception was hsa-miR-365b-5p, whose expression, analyzed by RT-qPCR in 5 BM groups, did not entirely correlate with the results from the differential expression analysis of small RNA sequencing data. However, since the Boruta algorithm identified it as part of the collapsed 6-miRNA signature, its expression changes across the 5 groups must have added a significant value to the model. After testing the 6-miRNA model on the RT-qPCR dataset, the sensitivity values were considerably high for all BM groups (>72%) except for BML (44.44%). An explanation may lie in the histological types of lung carcinomas, from which BML are derived. Lung carcinomas are subclassified into small cell lung carcinomas (SCLC) and non-small cell lung carcinomas (NSCLC), with the latter further divided into adenocarcinomas, squamous cell carcinomas, large cell carcinomas, and large cell neuroendocrine carcinomas (28). While the first treatment option for early stages of NSCLC is surgery and chemotherapy, radiotherapy, and immunotherapy for later NSCLC stages, the surgery in the case of SCLC is very limited and the first line of therapy is usually radiotherapy and chemotherapy (29,30). These differences in the management of patients with lung carcinomas are due to the high genetic heterogeneity of these tumors which are also present in BML and may have influenced the sensitivity of our classifier regarding BML samples. The specificity values were considerably high for all BM groups (>89%).
A few studies in the past have attempted to establish classifier models with the ability to classify tumor tissue samples, including metastatic tissues, according to their origin and miRNA expression using various approaches. Ferracin et al. (31) used a microarray platform to analyze the expression of 723 miRNAs in the training set of 40 primary tumors representing 10 different cancer types, validated their results in the test set of 45 primary tumor samples and 16 metastatic samples, and identified 47 differentially expressed miRNAs with the ability to correctly classify 100% of primary tumors and 78% of metastases. They found a high similarity of miRNA expression patterns in primary tumors and corresponding metastatic tumors, concluding that the primary origin appears to be the main determinant of the metastasis miRNA profile. They also found an exclusive miRNA expression pattern for every cancer type except lung and breast carcinomas, which exhibited similar expression profiles (31). This is consistent with our own findings since we also found noticeable overlap in the case of BML and BMB samples after the unsupervised clustering of BMs based on 58 differentially expressed miRNAs (Figure 1A), and in the results from the validation of our classifier algorithm which misclassified 10 BML and 4 BMB samples as BMB and BML, respectively (Figure 4). Ferracin et al. also observed high expression of hsa-miR-211-5p, hsa-miR-146a-5p and hsa-miR-514 in melanomas, hsa-miR-122-5p in renal carcinomas, and hsa-miR-192-5p, hsa-miR-194-5p and hsa-miR-215-5p in colon cancer (31), which agrees with our results (Figure 1). A study by Rosenfeld et al. (32) described a decision-tree classification algorithm based on the data from microarray expression profiling of more than 600 miRNAs in 205 primary tumors and 131 metastatic tumors, representing 22 different tumor origins. Even though some differences in miRNA expression between primary and metastatic tumors may be expected, the team reported none in the case of most cancers, including breast and colon tumors, concluding that primary tumors are suitable for training a classifier for metastases. They then cross-validated their decision tree within the training sample set, confirmed classifier performance on an independent blinded 83-sample test set and validated miRNA expression using RT-qPCR in additional 65 samples; they found that its classification accuracy reached 100% for most tissue classes, including all metastatic tumors. Although the nodes in their decision-tree classifier pertaining to lung, breast, renal and colorectal carcinomas and melanomas were based on the expression of different miRNAs than the ones we identified as the most differentially expressed when comparing one group of BMs with another group composed of other BMs, they also observed that the most common error was misclassification as lung cancer, similarly to our own observation regarding our classifier model, which both misclassified BML as BMs of different origin and other BMs as BML. The team also found the expression of miR-200 family to be the basis of the first major division, separating tissues of epithelial origin, e.g., lung, breast, and colon cancer, from tissues of other or mixed origin, e.g., melanomas and kidneys (32). This agrees with our study, which identified all 9 members of the family as ones of the most differentially expressed miRNAs among the 5 groups of BMs (Figure 1A) and as a part of our initial 32-miRNA classifier model. Rosenwald et al. (33) then improved the classifier from the previous study (32) by expanding the microarray dataset by profiling 254 additional FFPE tumor samples on a different, custom-designed microarray platform and by further testing this classifier in an expanded training set of 356 samples and validating it in an independent cohort of 204 samples. For 124 samples, the two algorithms, which the classifier is based on, generated a consensus prediction for a single tissue of origin. Out of these samples, 52 belonged to the 5 tissue types, from which BMs most frequently originate. The lowest sensitivity was observed for breast cancer (53%) and the highest for lung cancer (95%), while for colorectal and renal clear cell carcinomas and melanomas, the sensitivity was 80% or more. The specificity was higher than 94% for all 5 tumor types. Interestingly, 6 out of the 13 misclassified samples, which in reality were of breast, kidney or melanoma origin, were classified as lung tissue (33). These results again mostly agree with our findings with notable exception being the sensitivity for tumors with lung origin since the sensitivity in our case was much lower (44%). Varadhachary et al. (34) then prospectively evaluated the clinical utility of the improved classifier from the previous study (33) for patients with cancer of unknown primary (CUP) in the context of working diagnoses, which were predicted based on the available immunohistochemistry and clinicopathological data. The authors reported that 62 out of the 74 successfully profiled samples with unknown origin had their miRNA profile consistent or compatible with the clinical and/or pathological presentation. They also observed the improved classifier to be consistent in identifying cases where the most likely diagnosis is colon adenocarcinoma (34). Meiri et al. (35) then took this classifier and developed a second-generation diagnostic assay to identify a wider range of tumor types based on the expression of 64 miRNAs. They validated the assay on 509 blinded FFPE tissue samples of known origin and classified them into 42 tumor types, demonstrating an overall assay sensitivity of 85% and sensitivity of 90% for 403 cases with single tissue of origin predicted. Regarding the single-answer cases with predicted origin in the five tissue types, from which most BMs are derived, the sensitivity was lowest for breast cancer (75%) and highest for lung carcinomas and melanomas (100%) (35), which is consistent with the findings of the previous study (33) but again differs in terms of lung carcinomas from our observation. Meiri et al. (35) then applied the improved classifier on 52 CUP cases, reporting a prediction of convincing suggested origin in 88% of cases, with single predicted diagnosis in 48% of cases. Their classifier was able to correctly predict the tissue of origin in most cases of suspected lung carcinomas, few cases of breast, gastrointestinal and renal clear cell carcinomas, and one case of skin melanoma (35). Laprovitera et al. (36) also focused on classifying CUP cases but decided to use droplet digital PCR. In their two-algorithm classifier model, they implemented two miRNA sets from previously mentioned studies (31,32) and expanded the miRNA signature with 10 additional miRNAs, which served as reference candidates or markers of novel tumor classes or histological types (36). They trained their classifier on a cohort of 96 primary tumors, comprising 16 different tumor types and 19 histological classes, which were chosen with focus on the most common origin sites of CUP. Using clustering analysis, the authors identified highly similar miRNA expression patterns to our own findings, specifically high expression of hsa-miR-122-5p and hsa-miR-194-5p in renal clear cell carcinoma samples; hsa-miR-146a-5p, hsa-miR-211-5p and hsa-miR-514a-3p in melanoma samples; hsa-miR-194-5p, hsa-miR-200b-3p and hsa-miR-215-5p in colorectal carcinoma samples; and hsa-miR-141-3p and hsa-miR-200b-3p in triple-negative breast carcinomas and certain subtypes of lung carcinomas. They then evaluated the droplet digital PCR data using their classifier and reported generally low error rates for colorectal carcinoma, melanoma, and lung adenocarcinoma samples (16% or smaller) and higher error rates for renal tumor samples (25% or higher) and triple-negative breast carcinoma samples (53% or higher). Finally, the authors used the classifier for the prediction of tissue origin of 53 CUP cases and observed agreement between the two classifier algorithms in 94% of cases and compatibility of the prediction with clinicopathological diagnosis in 53% of cases. All mentioned studies demonstrate that miRNA-based classifiers could potentially serve as very useful tools in predicting tissue origin for CUP cases and, after further training and testing, possibly also for BMUP patients. One limitation of these studies, however, was the use of a pre-selected microarray panel of miRNAs, which, unlike small RNA sequencing, does not allow the exploration of the diagnostic potential of all known human miRNAs.
Conclusion
To our current knowledge, our study is the first to analyze miRNA expression in the five most frequent types of BMs using small RNA sequencing. The correct diagnosis or subtype of a tumor is crucial for further postoperative treatment, which will be even more accentuated in the future due to the increasing number of drugs indicated only for certain diagnoses. Correct diagnosis is also essential for valid estimation of patient prognosis, as highlighted in numerous published papers evaluating clinical prognostic markers. The currently most widely used assessment in routine clinical practice, the disease-specific Graded Prognostic Assessment (DS-GPA) (2), involves an online tool for valid estimation of prognosis. However, the tool is based on known primary cancer, making the correct diagnosis even more important. Our findings prove that miRNAs are significantly differentially expressed in BMs and that their expression patterns are specific for all investigated histological types of BM except for BML, which may require subdividing in future studies to reflect the differences in their genetic background and the resulting management of tumors with this origin. All in all, our study confirms the potential of miRNAs to serve as clinically relevant diagnostic biomarkers.
Conflicts of Interest
The Authors declare that they have no conflicts of interest regarding this study.
Authors’ Contributions
JS, MS, and RJ conceptualized the study and acquired funding. RJ, MHer, MS, and OS supervised the study. JS took care of the project administration. IR, JS, and OS devised the methodology used in the study. RJ, MHer, MS, LK, and PK provided the patient samples. IR, MV, LR, FS, and KT performed experimental investigation, including RNA isolation, quality and quantity control, library preparation, small RNA sequencing, bioinformatics processing of measured data, statistical analyses, and PCR validation. MHen, TK, VV, KT, IK, and HV curated all clinicopathological and experimental data. LR, MV, and JS performed the data visualization. IR, MV, and LR wrote the original draft. JS, OS, and TK revised the original draft. All Authors read and approved the manuscript.
Acknowledgements
This work was financially supported by the Ministry of Health of the Czech Republic (grant project nr. NV18-03-00398), by the Ministry of Education, Youth, and Sports of the Czech Republic (program EXCELES; grant projects nr. LX22NPO5102 and LX22NPO5107; financed by the European Union – Next Generation EU), and conceptual development of research organizations (Masaryk Memorial Cancer Institute, 00209805; University Hospital Brno, 65269705). The funders had no role in the design of the study, the collection, analysis, and interpretation of data, and in writing of the manuscript. We gratefully acknowledge the Core Facility Genomics, supported by the NCMG research infrastructure (project LM2018132 funded by MEYS CR), and Core Facility Bioinformatics of CEITEC Masaryk University for their support with obtaining of the scientific data presented in this paper.
References
- 1.Ostrom QT, Wright CH, Barnholtz-Sloan JS. Brain metastases: epidemiology. Handb Clin Neurol. 2018;149:27–42. doi: 10.1016/B978-0-12-811161-1.00002-5. [DOI] [PubMed] [Google Scholar]
- 2.Sperduto PW, Mesko S, Li J, Cagney D, Aizer A, Lin NU, Nesbit E, Kruser TJ, Chan J, Braunstein S, Lee J, Kirkpatrick JP, Breen W, Brown PD, Shi D, Shih HA, Soliman H, Sahgal A, Shanley R, Sperduto WA, Lou E, Everett A, Boggs DH, Masucci L, Roberge D, Remick J, Plichta K, Buatti JM, Jain S, Gaspar LE, Wu CC, Wang TJC, Bryant J, Chuong M, An Y, Chiang V, Nakano T, Aoyama H, Mehta MP. Survival in patients with brain metastases: summary report on the updated diagnosis-specific graded prognostic assessment and definition of the eligibility quotient. J Clin Oncol. 2020;38(32):3773–3784. doi: 10.1200/JCO.20.01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pentheroudakis G, Golfinopoulos V, Pavlidis N. Switching benchmarks in cancer of unknown primary: from autopsy to microarray. Eur J Cancer. 2007;43(14):2026–2036. doi: 10.1016/j.ejca.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 4.Lamba N, Wen PY, Aizer AA. Epidemiology of brain metastases and leptomeningeal disease. Neuro Oncol. 2021;23(9):1447–1456. doi: 10.1093/neuonc/noab101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnholtz-Sloan JS, Sloan AE, Davis FG, Vigneau FD, Lai P, Sawaya RE. Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the Metropolitan Detroit Cancer Surveillance System. J Clin Oncol. 2004;22(14):2865–2872. doi: 10.1200/JCO.2004.12.149. [DOI] [PubMed] [Google Scholar]
- 6.Berghoff AS, Schur S, Füreder LM, Gatterbauer B, Dieckmann K, Widhalm G, Hainfellner J, Zielinski CC, Birner P, Bartsch R, Preusser M. Descriptive statistical analysis of a real life cohort of 2419 patients with brain metastases of solid cancers. ESMO Open. 2016;1(2):e000024. doi: 10.1136/esmoopen-2015-000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nayak L, Lee EQ, Wen PY. Epidemiology of brain metastases. Curr Oncol Rep. 2012;14(1):48–54. doi: 10.1007/s11912-011-0203-y. [DOI] [PubMed] [Google Scholar]
- 8.Sperduto PW, Chao ST, Sneed PK, Luo X, Suh J, Roberge D, Bhatt A, Jensen AW, Brown PD, Shih H, Kirkpatrick J, Schwer A, Gaspar LE, Fiveash JB, Chiang V, Knisely J, Sperduto CM, Mehta M. Diagnosis-specific prognostic factors, indexes, and treatment outcomes for patients with newly diagnosed brain metastases: a multi-institutional analysis of 4,259 patients. Int J Radiat Oncol Biol Phys. 2010;77(3):655–661. doi: 10.1016/j.ijrobp.2009.08.025. [DOI] [PubMed] [Google Scholar]
- 9.Nieder C, Spanne O, Mehta MP, Grosu AL, Geinitz H. Presentation, patterns of care, and survival in patients with brain metastases: what has changed in the last 20 years. Cancer. 2011;117(11):2505–2512. doi: 10.1002/cncr.25707. [DOI] [PubMed] [Google Scholar]
- 10.Balestrino R, Rudà R, Soffietti R. Brain metastasis from unknown primary tumour: moving from old retrospective studies to clinical trials on targeted agents. Cancers (Basel) 2020;12(11):3350. doi: 10.3390/cancers12113350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavlidis N, Pentheroudakis G. Cancer of unknown primary site. Lancet. 2012;379(9824):1428–1435. doi: 10.1016/S0140-6736(11)61178-1. [DOI] [PubMed] [Google Scholar]
- 12.Kakimoto Y, Tanaka M, Kamiguchi H, Ochiai E, Osawa M. MicroRNA stability in FFPE tissue samples: Dependence on GC content. PLoS One. 2016;11(9):e0163125. doi: 10.1371/journal.pone.0163125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siegl F, Vecera M, Roskova I, Smrcka M, Jancalek R, Kazda T, Slaby O, Sana J. The significance of microRNAs in the molecular pathology of brain metastases. Cancers (Basel) 2022;14(14):3386. doi: 10.3390/cancers14143386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Illumina: NextSeq System Denature and Dilute Libraries Guide. Available at: https://support.illumina.com/downloads/nextseq-500-denaturing-diluting-libraries-15048776.html. [Last accessed on October 25, 2022]
- 15.Andrews S. FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. [Last accessed on October 25, 2022]
- 16.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2014;17(1):10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 17.Hannon GJ. FASTX-Toolkit. . Available at: http://hannonlab.cshl.edu/fastx_toolkit. [Last accessed on October 25, 2022]
- 18.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pantano L, Estivill X, Martí E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res. 2010;38(5):e34. doi: 10.1093/nar/gkp1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kursa M, Rudnicki W. Feature selection with the Boruta package. Journal of Statistical Software. 2015;36(11):1–13. doi: 10.18637/jss.v036.i11. [DOI] [Google Scholar]
- 23.Bonneau E, Neveu B, Kostantin E, Tsongalis GJ, De Guire V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC. 2019;30(2):114–127. [PMC free article] [PubMed] [Google Scholar]
- 24.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5(3):115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65(23):3756–3788. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283(22):14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, Martinvalet D, Song E, Lim B, Lieberman J. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One. 2009;4(9):e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholson AG, Tsao MS, Beasley MB, Borczuk AC, Brambilla E, Cooper WA, Dacic S, Jain D, Kerr KM, Lantuejoul S, Noguchi M, Papotti M, Rekhtman N, Scagliotti G, van Schil P, Sholl L, Yatabe Y, Yoshida A, Travis WD. The 2021 WHO classification of lung tumors: Impact of advances since 2015. J Thorac Oncol. 2022;17(3):362–387. doi: 10.1016/j.jtho.2021.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Alexander M, Kim SY, Cheng H. Update 2020: Management of non-small cell lung cancer. Lung. 2020;198(6):897–907. doi: 10.1007/s00408-020-00407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiddinga BI, Raskin J, Janssens A, Pauwels P, Van Meerbeeck JP. Recent developments in the treatment of small cell lung cancer. Eur Respir Rev. 2021;30(161):210079. doi: 10.1183/16000617.0079-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferracin M, Pedriali M, Veronese A, Zagatti B, Gafà R, Magri E, Lunardi M, Munerato G, Querzoli G, Maestri I, Ulazzi L, Nenci I, Croce CM, Lanza G, Querzoli P, Negrini M. MicroRNA profiling for the identification of cancers with unknown primary tissue-of-origin. J Pathol. 2011;225(1):43–53. doi: 10.1002/path.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, Lebanony D, Goren Y, Silberschein E, Targan N, Ben-Ari A, Gilad S, Sion-Vardy N, Tobar A, Feinmesser M, Kharenko O, Nativ O, Nass D, Perelman M, Yosepovich A, Shalmon B, Polak-Charcon S, Fridman E, Avniel A, Bentwich I, Bentwich Z, Cohen D, Chajut A, Barshack I. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26(4):462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 33.Rosenwald S, Gilad S, Benjamin S, Lebanony D, Dromi N, Faerman A, Benjamin H, Tamir R, Ezagouri M, Goren E, Barshack I, Nass D, Tobar A, Feinmesser M, Rosenfeld N, Leizerman I, Ashkenazi K, Spector Y, Chajut A, Aharonov R. Validation of a microRNA-based qRT-PCR test for accurate identification of tumor tissue origin. Mod Pathol. 2010;23(6):814–823. doi: 10.1038/modpathol.2010.57. [DOI] [PubMed] [Google Scholar]
- 34.Varadhachary GR, Spector Y, Abbruzzese JL, Rosenwald S, Wang H, Aharonov R, Carlson HR, Cohen D, Karanth S, Macinskas J, Lenzi R, Chajut A, Edmonston TB, Raber MN. Prospective gene signature study using microRNA to identify the tissue of origin in patients with carcinoma of unknown primary. Clin Cancer Res. 2011;17(12):4063–4070. doi: 10.1158/1078-0432.CCR-10-2599. [DOI] [PubMed] [Google Scholar]
- 35.Meiri E, Mueller WC, Rosenwald S, Zepeniuk M, Klinke E, Edmonston TB, Werner M, Lass U, Barshack I, Feinmesser M, Huszar M, Fogt F, Ashkenazi K, Sanden M, Goren E, Dromi N, Zion O, Burnstein I, Chajut A, Spector Y, Aharonov R. A second-generation microRNA-based assay for diagnosing tumor tissue origin. Oncologist. 2012;17(6):801–812. doi: 10.1634/theoncologist.2011-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laprovitera N, Riefolo M, Porcellini E, Durante G, Garajova I, Vasuri F, Aigelsreiter A, Dandachi N, Benvenuto G, Agostinis F, Sabbioni S, Berindan Neagoe I, Romualdi C, Ardizzoni A, Trerè D, Pichler M, D’Errico A, Ferracin M. MicroRNA expression profiling with a droplet digital PCR assay enables molecular diagnosis and prognosis of cancers of unknown primary. Mol Oncol. 2021;15(10):2732–2751. doi: 10.1002/1878-0261.13026. [DOI] [PMC free article] [PubMed] [Google Scholar]