Abstract
Hypothalamic gonadotropin-releasing hormone (GnRH) neurons lay the foundation for human development and reproduction; however, the critical cell populations and the entangled mechanisms underlying the development of human GnRH neurons remain poorly understood. Here, by using our established human pluripotent stem cell-derived GnRH neuron model, we decoded the cellular heterogeneity and differentiation trajectories at the single-cell level. We found that a glutamatergic neuron population, which generated together with GnRH neurons, showed similar transcriptomic properties with olfactory sensory neuron and provided the migratory path for GnRH neurons. Through trajectory analysis, we identified a specific gene module activated along the GnRH neuron differentiation lineage, and we examined one of the transcription factors, DLX5, expression in human fetal GnRH neurons. Furthermore, we found that Wnt inhibition could increase DLX5 expression and improve the GnRH neuron differentiation efficiency through promoting neurogenesis and switching the differentiation fates of neural progenitors into glutamatergic neurons/GnRH neurons. Our research comprehensively reveals the dynamic cell population transition and gene regulatory network during GnRH neuron differentiation.
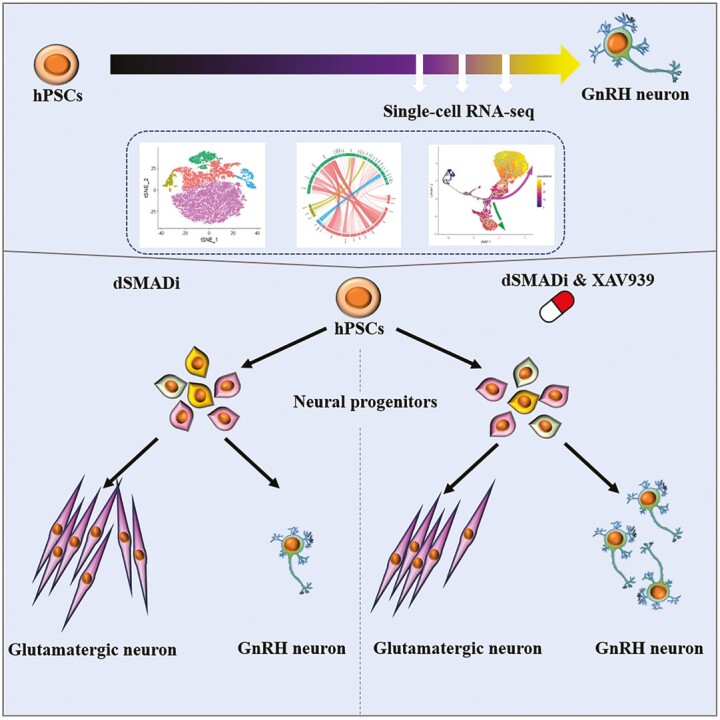
Graphical Abstract
Graphical Abstract.
Significance Statement.
This study first identified and characterized diverse cell types generated along the GnRH neuron differentiation protocol. Additionally, by analyzing the differentiation trajectories, we improved the efficiency of GnRH neuron differentiation with Wnt signaling inhibition.
Introduction
Gonadotropin-releasing hormone (GnRH) neurons originate in the olfactory placode region and subsequently migrate from the nose to the brain along the terminal, vomeronasal, and olfactory nerves.1-4 GnRH neurons residing in the hypothalamus secrete GnRH decapeptide in the hypophyseal portal system, and thereby control the synthesis and release of pituitary gonadotropins, ie, luteinizing hormone (LH) and follicle-stimulating hormone (FSH), which then stimulate gonadal steroidogenesis and gametogenesis.5 Disruption of GnRH neuron development, GnRH signaling or GnRH decapeptide secretion can lead to congenital hypogonadotropic hypogonadism (CHH) featuring complete or partial pubertal failure and infertility. Until now, mutations in more than 60 genes have been implicated in CHH,6 and several genes, such as Dlx family members, Otx2, Pou2f1, and Msx1, have been identified as Gnrh promoter modulators in murine GnRH neuron models.7,8 Despite the progress in this area, information about how gene-environment interactions significantly influence GnRH neuron development in humans is still fragmentary.
Human pluripotent stem cells (hPSCs), which can self-renew indefinitely in culture while maintaining the ability to become any cell type in the human body,9,10 provide a possibility to model the development and function of human GnRH neurons in vitro. Accordingly, we have previously developed a protocol to differentiate hPSCs into GnRH decapeptide-secreting neurons with dual-SMAD inhibition (dSMADi), followed by FGF8 treatment and Notch inhibition.11 By now, we have used this model to investigate the role of MKRN3 in GnRH neuron differentiation12 and to describe transcriptome profiles of the putative GnRH neuron progenitors and early postmitotic GnRH neurons with bulk RNA sequencing.13 The efficiency of our protocol is, however, approximately 15%,11 which suggests that it generates multiple cell types. Single-cell RNA sequencing (scRNA-seq) can reveal the heterogeneity of cells, allow the identification of rare cell populations, uncover putative regulatory relationships between genes, and track the developmental trajectories of distinct cell lineages during their differentiation.14
Here, we aimed to identify the developmental trajectories that lead to GnRH neuron fate in vitro using scRNA-seq. We defined emerging cell types along the differentiation process based on their global gene-expression profiles to create a precise cell-by-cell description of in vitro GnRH neuron differentiation and reconstructed the developmental trajectory of GnRH neuron specification. Based on the development trajectory, we identified 443 putative factors involved in the patterning of GnRH neuron differentiation and examined the expression of DLX5, one of the most noticeable DLX family genes, in human GnRH neurons both in vitro and in vivo. Furthermore, we observed that Wnt signaling is downregulated along the GnRH neuron specification. Given that Wnts could play a role in murine frontonasal development,15,16 and Wnt signaling regulates neural progenitors (NPs) heterogeneity and specifies their regional identity,17 we investigated the Wnt effect on the GnRH neuron differentiation by applying XAV939, a Wnt inhibitor, during dSMADi phase. Remarkably, we found that Wnt inhibition affects the cell fate determination already during the first 10 days, and significantly improved the yield of hPSC-derived GnRH neurons.
Together, the identified transcription factors and the role of Wnt signaling as a determinant of GnRH neuron fate patterning have potential implications for mechanisms of puberty, and drug development of contraception and hormone-dependent cancer.
Materials and Methods
Stem Cell Maintaining and Differentiation
Human embryonic stem cell line (WA09) and 2 hiPSC lines HEL11.4 and HEL24.3 were used in this study.
Human pluripotent stem cells were grown and expanded in monolayer on plastic cell culture dishes (Corning) coated with Matrigel (CORNING, Cat. 356231) in mTeSR1 (STEMCELL, Cat. 85850) cell culture medium, and dissociated for replating using 0.5 mM Ultrapure EDTA (15575-038, Invitrogen) in PBS (Corning) every 5 days. Before the experiment started, the cells were grown until approximately 95% confluence, covering most of the surface area of the dish. GnRH neuron differentiation was performed as described before.11 In brief, N2B27 medium (Neurobasal medium supplemented with N2 and B27 supplement, in 1:1 ratio with DMEM/F12, and containing 1× Glutamax (ThermoFisher Scientific), 100 U/mL penicillin, and 100 µg/mL streptomycin (Sigma)) was supplemented with 2 µM dorsomorphin (Selleckchem, Cat. S7306) and 10 µM SB431542 (Sigma, Cat. S4317), and replaced daily during the first 10 days. On day10, cells were replated in 1:2 ratio using 200 U/mL Collagenase IV (ThermoFisher Scientific) onto Matrigel-coated dishes in N2B27 supplemented with 10 µM Y-27632 (Selleckchem, Cat. S1049). From day 11 to day 20, N2B27 was supplemented with 100 ng/mL of FGF8 (PeproTech, Cat. AF-100-25). Cell vials were frozen for storage on day 17 by dissociation with EDTA and resuspension in a freezing medium containing N2B27, 10% DMSO HybriMax (D2650, Sigma-Aldrich) and 10 µM Y-27632 in 2 mL cryovial tubes (ThermoFisher Scientific) stored at −150 °C. Cells were thawed in N2B27 containing 100 ng/mL FGF8 and 10 µM Y-27632, and grown until they were confluent (approximately 3 days). On day 20, cells were detached using 0.5 mM EDTA and replated in N2B27 at 1:8 ratio. The next day following the split, N2B27 was supplemented with 20 µM DAPT (Selleckchem, Cat. S2215) and the medium was replaced every 2 days. For Wnt inhibition, based on the original differentiation protocol, we added 10 µM XAV939 (Selleckchem, Cat. S1180) from day 0 to day 10.
Single-cell Isolation
Samples from days 22, 24, and 26: Medium was removed and cells were washed one time with DMEM/F12 and one time with sterile PBS, and incubated in 2 mg/mL Papain solution (STEMCELL, Cat. 07465) for 7 min at 37 °C. An equal volume of 1% BSA in PBS without Ca2+ and Mg2+ was added to the cell suspension, and cells were suspended by pipetting up and down approximately 15 times using a 1-mL pipette tip. The suspension was moved to a 2 mL Eppendorf tube and centrifuged at 300 × g for 3 min. The supernatant was removed and cells were resuspended in 1% BSA + 2 µM EDTA in PBS without Ca2+ and Mg2+, where volume was adjusted based on a viable cell count of approximately 1000 cells/µL using LUNA-FL Dual Fluorescence Cell Counter. The cell suspension was filtered 2-4 times through a 35-µm cell strainer tube cap (Falcon, Cat. 352235), and processed for scRNA-seq library construction and sequencing.
Samples from day 0, day 5, and day 10: medium was removed and cells were washed one time with sterile PBS and incubated in Accutase (ThermoFisher, Cat. A1110501) for 4 min at 37 °C. An equal volume of 1% BSA in PBS without Ca2+ and Mg2+ was added to the cell suspension, and cells were suspended by pipetting up and down approximately 15 times using a 1-mL pipette tip. The suspension was moved to a 2-mL Eppendorf tube and centrifuged at 300 × g for 3 min. The supernatant was removed and cells were resuspended in 1% BSA + 2 µM EDTA in PBS without Ca2+ and Mg2+. Cell numbers were counted using LUNA-FLTM Dual Fluorescence Cell Counter for further labeling.
Cell Multiplexing Oligo Labeling for Single-cell RNA Sequencing
Five samples (D0, D5_dSMADi, D5_XAV939, D10_dSMADi, and D10_XAV939) were labeled with cell multiplexing oligo according to the manufacturer’s instructions (10× Genomics, 3ʹ CellPlex Kit Set A, Cat. 1000261). Briefly, 2 × 106 cells from each sample were suspended in 100 µL Cell multiplexing Oligo (301–305), and incubated for 5 minutes at room temperature. The samples were washed by Wash & Resuspension Buffer 3 times. The cells were counted and then mixed as calculated for scRNA-seq library construction.
scRNA-seq Library Construction and Sequencing
Single-cell RNA-sequencing was performed using the chromium single-cell 3ʹ RNA-sequencing system (10× Genomics, Pleasanton, CA) with the Reagent Kit v3 according to the manufacturer’s instructions. Briefly, the cells were loaded into chromium single cell B chip (10× Genomics, Pleasanton, CA) and gel beads in emulsion (GEM) generation were performed aiming at 10 000 cell capture per sample. Subsequent cDNA purification, amplification, and library construction were performed as instructed. Sample libraries were sequenced on an Illumina NovaSeq 6000 system using following read lengths: 28 bp (Read 1), 8 bp (i7 Index), 0 bp (i5 Index), and 89 bp (Read 2) aiming at 50 000 reads per cell sequencing depth.
The Cell Ranger v3.1 mkfastq and count pipelines (10× Genomics, Pleasanton, CA) were used to demultiplex and convert chromium single-cell 3ʹ RNA-sequencing barcodes and read data to FASTQ files and to generate aligned reads and gene-cell matrices. Reads were aligned to the human genome GRCh38.
Processing the scRNA-seq Data
All main processing steps were performed with Seurat v.3.1. Quality control was first performed independently on each library to find appropriate filtering thresholds (see Supplementary Table S6 for detailed parameters of each library). Expression matrices for each sample were loaded into R as Seurat objects, retaining only cells that fit the standards. Genes significantly enriched in each cluster were identified using the default algorithm in Seurat.18,19 Functional annotation of the resulting marker genes list related to GO terms were performed using g:Profiler (https://biit.cs.ut.ee/gprofiler/gost). Intercellular communication is modeled by the NicheNet package.20 Trajectory analysis of the GnRH neuron development was performed using Monocle (v3).21,22 A comparison of the similarity between the Glut cluster identified from our scRNA-seq data and different cell types identified in the reference transcriptomic datasets (olfactory cleft and turbinate tissue scRNA-seq datasets) was performed using SingleR.23
Immunofluorescence Assays
The cells grown on the Matrigel coated coverslips were fixed in 4% paraformaldehyde for 15 minutes at room temperature. Triton X-100 (0.25%) was used for permeabilization, followed by blocking reagent (ThermoFisher Scientific, Cat. B10710). Samples were incubated with primary antibodies (Supplementary Table S4) overnight at 4 °C, and then with secondary antibodies for 2 h at room temperature. DAPI was used to counterstain nuclei. After the samples were mounted, they were imaged by using a widefield microscope (Nikon Eclipse Ti-E, Japan) or a confocal microscope (Leica TCS SP8, Japan).
Human Fetuses
Fetal tissues were made available in accordance with French bylaws (Good Practice Concerning the Conservation, Transformation, and Transportation of Human Tissue to Be Used Therapeutically, published on December 29, 1998). The studies on human fetal tissue were approved by the French agency for biomedical research (Agence de la Biome´ decine, Saint-Denis la Plaine, France, protocol n°: PFS16-002). Non-pathological human fetuses were obtained at GW8.5 and GW13 from pregnancies terminated voluntarily after written informed consent of the parents (Gynaecology Department, Jeanne de Flandre Hospital, Lille, France).
Multiplex Fluorescence In Situ Hybridization (FISH) Combined With Immunofluorescence
Two human fetuses (males; 8.5 and 13 gestational weeks post-amenorrhea; GW) were obtained from voluntarily terminated pregnancy with the parent’s written informed consent (Gynecology Hospital Jean de Flandre, Lille, France). The fetuses were snap-frozen in liquid nitrogen and stored at −80 °C until use. Human tissues were cryosectioned using a CM3050 Leica Cryostat at 16 μm.
FISH was performed on frozen sections of the nasal region of a GW 8.5 and a GW 13 fetus with the RNAscope Multiplex Fluorescent Kit v2 according to the manufacturer’s protocol (Advanced Cell Diagnostics). Specific probe was used to detect DLX5 (Hs-DLX5-C3 REF 569471-C3) mRNA. Hybridization with a probe against the Bacillus subtilis dihydrodipicolinate reductase (dapB) gene (320871) was used as a negative control. Immunofluorescence against GnRH was performed, after RNAscope staining, as previously reported.24 Briefly, the sections were rinsed with 0.1 M PBS and incubated at 4 °C overnight with the previously validated3 guinea-pig anti-GnRH (a generous gift from Dr. Erik Hrabovszky, Laboratory of Endocrine Neurobiology, Institute of Experimental Medicine of the Hungarian Academy of Sciences, Budapest, Hungary) diluted at 1:1000 in 0.1 M PBS containing 0.3% Triton X-100 and 10% normal donkey serum. The sections were then washed in PBS and incubated for 1 h with AlexaFluor 488-conjugated secondary antibody (Molecular Probes, Invitrogen, San Diego, CA) diluted 1:400 in PBS and counterstained with DAPI nuclear staining (1:10 000; Molecular Probes, Invitrogen, San Diego, CA). Sections were mounted using Mowiol (Calbiochem, USA) and analyzed using an LSM 710 confocal microscope (Zeiss).
Flow cytometry
The cells were dissociated into single cells and suspended in FACS buffer (10% (v/v) FBS and 1 mM EDTA in 1× HBSS). Flow cytometry was performed using a Sony SH800z sorter at the Biomedicum Flow Cytometry Unit, University of Helsinki. Data analyses were performed with FlowJo software (v10; FlowJo LLC, Ashland, OR, USA).
Real-time PCR
Total RNA was extracted using NucleoSpin RNA Plus (MACHEREY-NAGEL, 740984). mRNA was reverse transcribed using an iScript cDNA Synthesis Kit (Bio-Rad, 170-8891). Real-time quantitative PCR was performed with cDNA using HOT FIREPol EvaGreen qPCR Mix Plus (Solis BioDyne) and LightCycler 480 (Roche) for 45 cycles of 95 °C for 15 s, 60 °C for 20 s and 72 °C for 20 s. mRNA expression was normalized to cyclophilin G (PPIG) and all the primers used are listed in Supplementary Table S5.
Cell Migration Assay
Cells on day 20 were spited and seeded into the Incucyte Imagelock 96-well plates. A total of 20 µM DAPT was given on day 21 and media change was done every other day. The plate was imaged using an IncuCyte S3 Live Cell Analysis System (Sartorius Corporation) from day 22 to day 26 at 1 h intervals.
Quantification of Secreted GnRH
Cell culture medium was collected concurrently with medium refreshments at indicated time points. Medium was centrifuged shortly (1200 × g for 10 minutes at 4 °C) and stored in −80 °C. Secreted GnRH decapeptide was quantified by competitive Fluorescent Enzyme Immunoassay (EIA, Phoenix Pharmaceuticals, Inc., FEK-040-02) according to manufacturer’s instructions. GnRH concentrations were determined from the standard curve fitted with 4-point parameter logistic nonlinear regression model.
Statistical Tests
The results are shown as means ± SD of data from at least 3 separate experiments. Statistical analyses were performed using GraphPad Prism 8 (Graphpad Software) and FlowJo 10.7 software (BD Biosciences). For the comparison between 2 groups, we used t-test analysis. For the comparisons among 3 groups (D0, D25-dSMADi, and D25-XAV939), we used one-way ANOVA analysis (Tukey’s multiple comparisons test). For the comparisons among 5 groups (D0, D5-dSMADi, D5-XAV939, D10-dSMADi, and D10-XAV939) and ELISA results, we used 2-way ANOVA analysis (Sidak’s multiple comparisons test). P values of .05 or less were considered statistically significant, as indicated by asterisks in the figures.
Results
Diverse Cell Populations Are Generated by the GnRH Neuron Differentiation Protocol
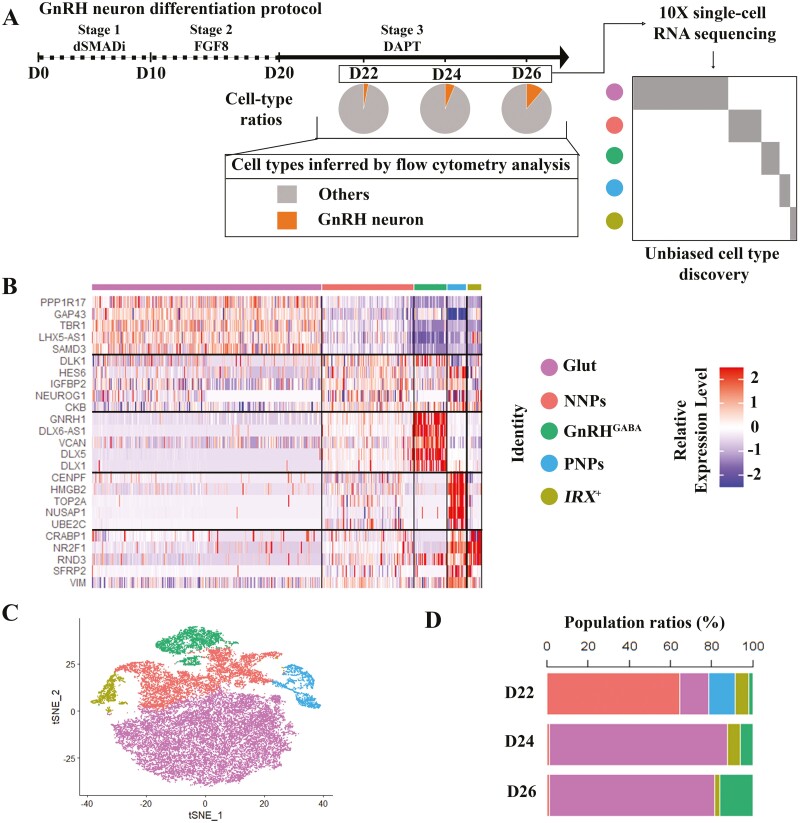
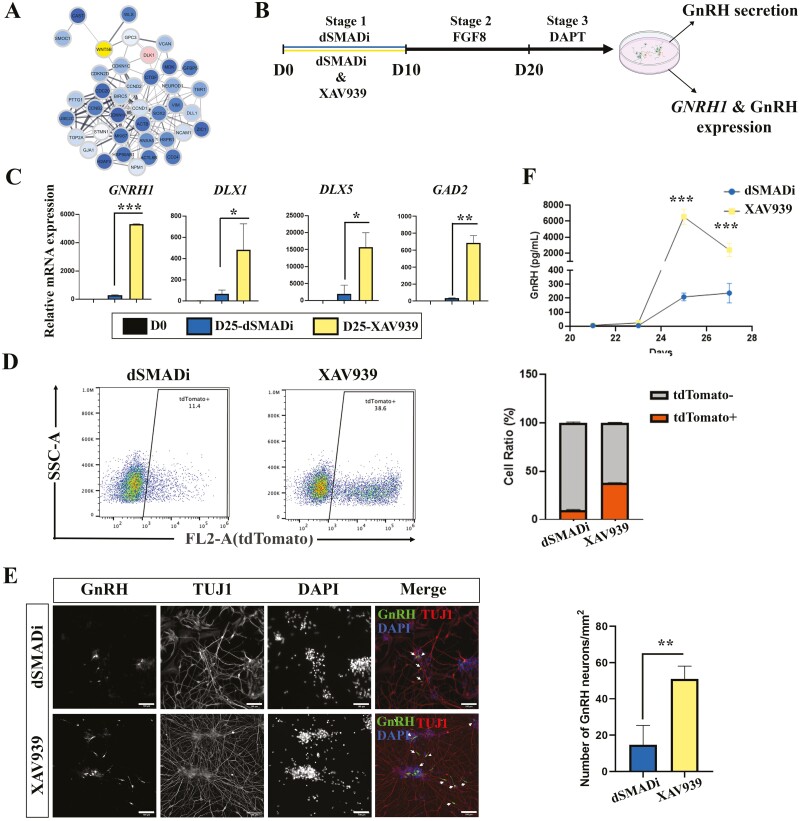
In the first phase of the GnRH neuron differentiation,11 dSMADi is used to produce NPs. This is followed by administration of FGF8 to pattern anterior neural progenitors and, finally, Notch inhibitor DAPT is used to induce neural maturation (Fig. 1A). We first investigated the temporal differentiation efficiency of GnRH neurons with flow cytometry analysis by using the GNRH1-tdTomato hPSC reporter line described before.13 The results showed that the cell population with tdTomato signal increased steadily from day 22 (D22, 3.2% ± 0.12%) to day 26 (D26, 10.9% ± 0.45%) (Fig. 1A and Supplementary Fig. S1).
Figure 1.
scRNA-seq reveals 5 cell clusters along GnRH neuron differentiation. (A) Schematic of GnRH neuron differentiation protocol and GnRH neuron population ratios identified by flow cytometry on day 22 (D22) (3.2 ± 0.12%), day 24 (D24) (6.3 ± 0.05%), and day 26 (D26) (10.9 ± 0.44%). (B) Heatmap of genes significantly enriched within each individual cluster. Single cells are shown in columns; genes are shown in rows. Top 5 differential genes of each cell cluster are shown. (C) t-SNE projections of cells sampled from all 3 days (D22, D24, and D26). Cells are colored according to their assigned cluster. (D) Components of different sample days (D22, D24, and D26) in each cluster during GnRH neuron differentiation. D = days of differentiation.
Next, we performed scRNA-seq of the cells collected on D22, D24, and D26 of differentiation with 10× Genomics (Fig. 1A), and recovered a total of 31 899 cells and 23 113 genes from all the samples (Supplementary Fig. S2). Following data processing, high-quality transcriptomic data were obtained from 29 686 cells, including 9523 cells from D22, 9107 cells from D24, and 11 056 cells from D26 (Supplementary Fig. S2). By performing Seurat analysis with transcriptomes of cells from D22, D24, and D26 separately, and also with integrated datasets (29 686 cells), we identified 5 distinct cell clusters during the differentiation process. These clusters were subsequently annotated based on the expression of known marker genes and an automatic annotation toolkit, scCATCH.25 The heatmap shows the expression of the top 5 marker genes across 5 clusters: (i) proliferating neural progenitors (PNPs) that express HMGB2, MKI67, and TOP2A that are markers for cell proliferation; (ii) nonproliferating neural progenitors (NNPs) that express DLK1, NEUROG1, NEUROD1/4, and SOX2 that most resembles neural progenitors; (iii) GnRH neurons (GnRHGABA) that express GNRH1, GAD2, and DLX family genes, such as DLX1, DLX2, and DLX5; (iv) glutamatergic neurons (Glut) that express PPP1R17, TBR1, and LHX family genes, such as LHX1, LHX2, LHX5, and LHX9; and (v) IRX+ which highly expresses CRABP1, NR2F1, and IRX family genes (Fig. 1B, Supplementary Table S1). We used t-distributed stochastic neighbor embedding (tSNE) to visualize the cell clusters (Fig. 1C, Supplementary Fig. S3). All 5 cell clusters were present on D22. However, the PNPs (D22: 13.0%) waned away on D24, and the proportion of NNPs also shrunk dramatically after D22. Conversely, the proportion of both Glut and GnRHGABA populations increased markedly along the differentiation (Fig. 1D). Thus, we identified 5 emergent cell types at the last stage of GnRH neuron differentiation.
Neural Progenitors Perform Different Functions and Have Different Differentiation Capacities
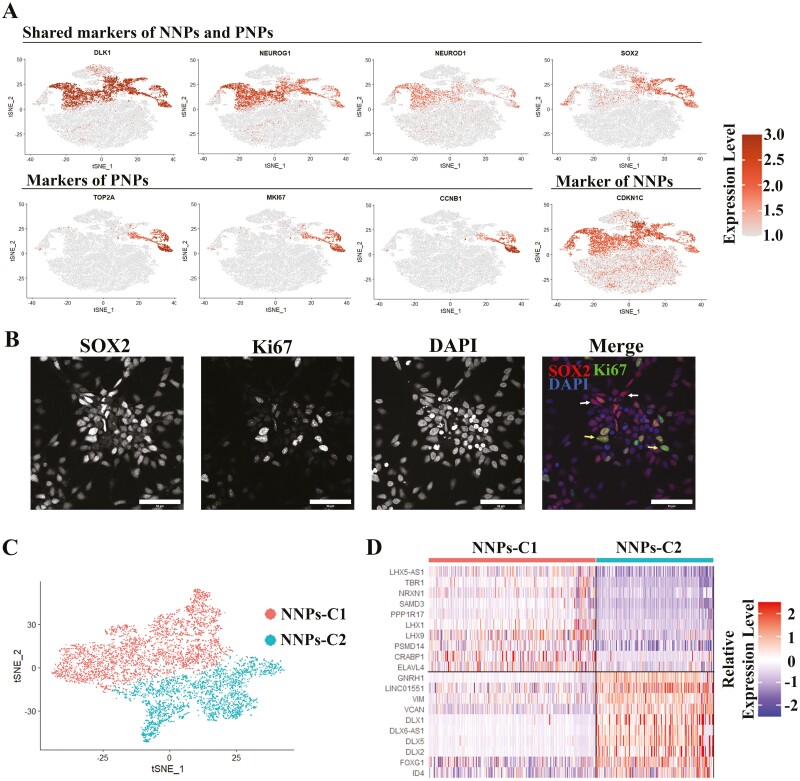
Based on the Seurat analysis, 2 types of NPs were identified, including PNPs and NNPs. Both expressed SOX2, DLK1, and basic helix-loop-helix family genes, including NEUROG1 and NEUROD1 (Fig. 2A) that have been reported to be expressed in neural progenitors in vivo.26-29 PNPs highly expressed cell-cycle-associated genes (eg, MKI67, TOP2A, CCNB1, and CCNB2), while the NNPs highly expressed STMN2, and CDKN1C, a cell-cycle-inhibitor gene (Fig. 2A and Supplementary Fig. S13). We applied Gene Ontology (GO) enrichment analysis based on the profiles of differentially expressed genes (DEGs) between these 2 types of NPs and identified that neuron differentiation and neurogenesis were enriched in NNPs, and cell division and mitotic cell cycle in PNPs (Supplementary Fig. S4A and S4B). Consistent with the gene expression profiles, immunostaining also verified the 2 types of NPs (SOX2+/Ki67+ and SOX2+/Ki67−) on D22 (Fig. 2B). Furthermore, we performed an unsupervised sub-clustering analysis of the cells in NNPs using Seurat, and we identified 2 sub-clusters from NNPs, named NNPs-C1 and NNPs-C2 (Fig. 2C). NNPs-C1 highly expresses PPP1R17, TBR1, LHX1, and LHX9, while the NNPs-C2 is marked by higher expression of GNRH1, DLX1, DLX2, and DLX5 (Fig. 2D and Supplementary Fig. S5). These genes are the markers for the Glut, and GnRHGABA cell clusters, respectively (Fig. 1B, Supplementary Table S1), which suggests that the NNPs have already committed to different lineages after exiting the cell cycle. Together, these data suggest that PNPs, which maintain the capacity of cell proliferation, are probably responsible for maintaining NPs pool, while NNPs have exit the pool of proliferative cells and exhibit differentiation capabilities for different neuronal lineages.
Figure 2.
Characterization of neural progenitors. (A) Expression of NNPs markers and PNPs markers projected onto the t-SNE map. (B) Immunofluorescence staining for 2 major neural progenitors (NNPs (SOX2+/Ki67−), and PNPs (SOX2+/Ki67+)). White arrows indicate NNPs, and yellow arrows indicate PNPs. Scale bar = 50 µm. (C) t-SNE projections of NNPs, which are further divided into 2 sub-clusters (NNPs-C1 and NNPs-C2). (D) Heatmap of differentially expressed genes between NNPs-C1 and NNPs-C2.
Glut Provides the Migratory Path for GnRHGABA and Displays Similar Transcriptional Properties with Olfactory Neurons
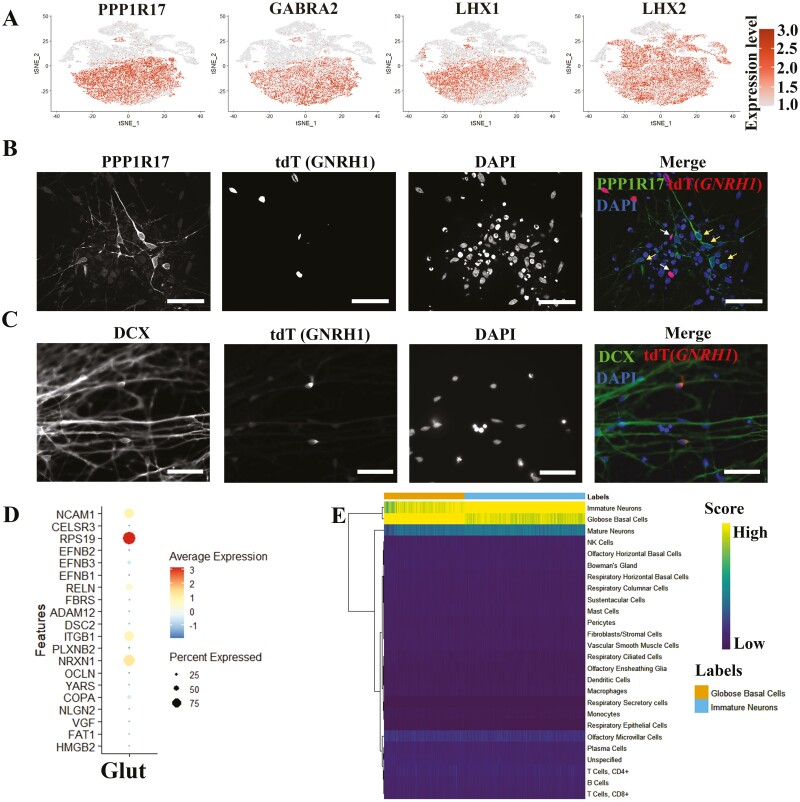
The cluster of Glut strongly expresses glutamatergic neuron markers, such as SLC17A6 (also named as VGLUT2), PPP1R17, GABRA2, LHX1, and LHX2 (Fig. 3A, Supplementary Table S1). GO enrichment analysis showed that neurogenesis, neuron differentiation, nervous system development, and cell projection morphogenesis were enriched in Glut cluster (Supplementary Fig. S4C). Through immunostaining of the differentiated GnRH reporter cell line (GNRH1-tdTomato hPSC, D25/D26), we verified that Glut (PPP1R17+), which are bipolar and show long axons, were non-GnRH neurons (tdTomato-) (Fig. 3B). Toward the end of the differentiation (D27), the neurons gathered together, and the PPP1R17+ cells (Glut) with extensively long axons formed the bundle of axons, with GnRH neurons set beside (Supplementary Fig. S6). To investigate the mobility of differentiated neurons, we took a video from D22 to D26 and noticed that the cultured cells organize in condensed nodes that are interconnected with bundles of axons. Notably, GnRH neurons (tdTomato+) were quite motile because they moved along the interconnecting tracts to integrate into the nodes (Supplementary Video S1). Next, we modeled the communications between Glut cluster and GnRHGABA cluster with Nichenet.20 The Glut cluster highly expressed genes such as NCAM1, RELN, ITGB1, and NRXN1 (Fig. 3D), and these genes regulate neuronal processes such as neuronal migration, and axon pathfinding,30-34 which suggests that the Glut may assist in the migration of GnRHGABA. Consistently, both Glut and GnRHGABA highly express DCX, which is a marker for immature neurons and migratory neurons (Fig. 3C).35,36 Given that axons of the olfactory neurons provide the early GnRH neuron migration pathway in vivo,37 we compared the Glut cluster with the published single-cell datasets about olfactory cleft and turbinate tissue,38 and found that immature neurons, globose basal cells, and mature neurons were the top 3 cell types according to the estimated similarity score. This indicates that Glut is highly similar with the immature olfactory neurons in regard to transcriptomic properties (Fig. 3E). By checking the specific markers, Glut cluster indeed highly expressed LHX2, EMX2, GNG8, and TUBB, markers of immature OSNs (Supplementary Fig. S7).38 Thus, we identified Glut population generated by GnRH neuron differentiation protocol, with possible correspondence to olfactory neurons.
Figure 3.
Characterization of glutamatergic neurons. (A) Expression of Glut markers projected onto the t-SNE map. (B) Immunofluorescence staining for Glut marker (PPP1R17) showing non co-localization with GnRH neurons. White arrows indicate GnRH neurons, and yellow arrows indicate Glut. Scale bar = 50 μm. (C) Immunofluorescence staining for migratory neuron marker (DCX) showing differentiated neurons are migratory. Scale bar = 50 µm. (D) Dot plot showing the expression level of prioritized ligands in Glut. (E) Comparing Glut cluster with published scRNA-seq dataset from olfactory cleft and turbinate tissues.
Identifying Differentiation Trajectory and Regulatory Genes During GnRH Neuron Differentiation Using Pseudotime Analysis
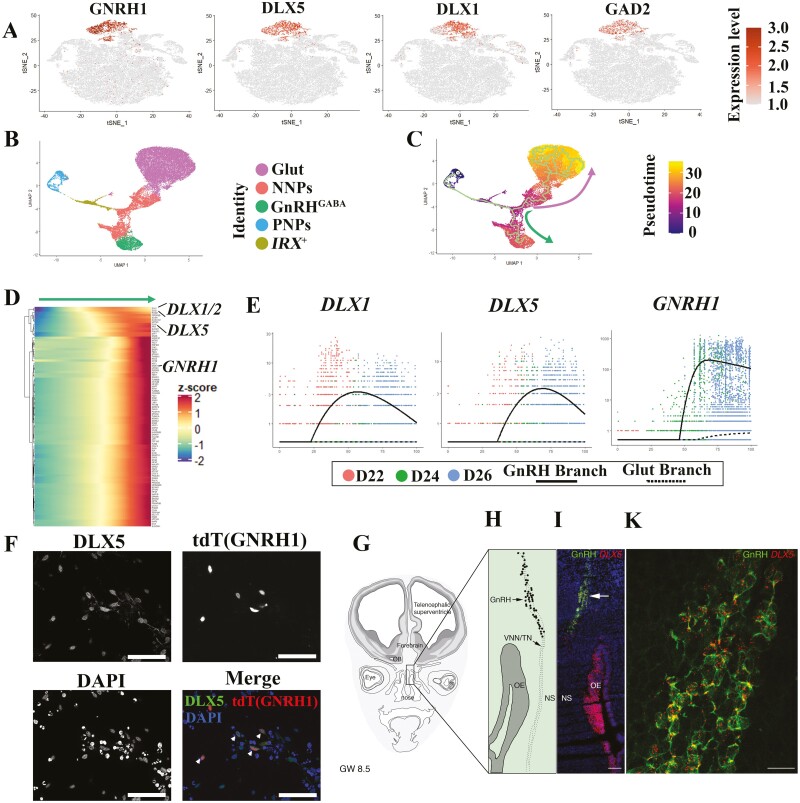
GnRH neurons were identified GABAergic and termed GnRHGABA. This cluster highly express GABAergic neuron markers, such as SLC32A1 (also named as VGAT), DLX1/2/5, and GAD2 (Fig. 4A, Supplementary Table S1) according to our analysis. We next sought to reveal the putative regulators pertinent to GnRH neuron development. We reconstructed the GnRH neuron differentiation trajectories by performing Monocle 3.22 After importing the integrated dataset from Seurat into Monocle 3, the cell distribution was displayed with Uniform Manifold Approximation and Projection (UMAP) (Fig. 4B). We found genes that vary in some interesting way across the clusters (Supplementary Fig. S8C and Table S2), and module 18 represents the co-regulated genes highly specific to GnRHGABA cluster. After building the trajectory, we noticed that there is a branch point during the differentiation, one path led by Glut lineage (upwards), and the other by GnRHGABA lineage (downwards) (Fig. 4C). Through analysis of each differentiation lineage, we identified the genes that vary across pseudotime (Fig. 4D, Supplementary Fig. S8A and S8B). Interestingly, in GnRHGABA lineage, DLX1/2 and DLX5 are activated before GNRH1 (Fig. 4D and 4E), and they are not activated in the Glut lineage (Fig. 4E). We observed that the differentiated GnRH neurons (tdTomato+) express stronger DLX5 than other cell types in the culture (Fig. 4F). To confirm DLX5 expression in human GnRH neurons in vivo, we performed RNAscope staining for DLX5 in human fetuses (8.5 gestational weeks) together with the expression of GnRH. Consistently, all GnRH-immunoreactive neurons expressed DLX5 throughout the migratory pathway (Fig. 4G-4K). In addition, the olfactory epithelium, which is postulated to be the origin of GnRH neurons,39 also highly expresses DLX5 (Fig. 4I). Together, these results model the trajectories for GnRH differentiation and indicate that the DLX family genes, including DLX5, are involved in GnRH neuron development.
Figure 4.
Reconstruction of the GnRH neuron differentiation trajectories. (A) Expression of GnRHGABA markers projected onto the t-SNE map. (B) UMAP projection of all cells with Monocle analysis. Cells are colored according to their assigned cluster. (C) UMAP shading of branch assignment and pseudotime value of each cell. Green arrow shows the GnRHGABA branch, and pink arrow shows the Glut branch. (D) Heatmap of differentially expressed genes (upregulated in GnRHGABA branch) identified by Monocle (rows), with cells (columns) ordered according to the pseudotime development. (E) Expression of selected marker genes along pseudotime ordering. Dots show expression in single cells. Lines show regression on pseudotime for each branch (solid line, GnRHGABA; dash line, Glut). (F) Immunofluorescence staining for GnRHGABA marker (DLX5) showing co-localization with GnRH neurons. Scale bar = 50 µm. Arrows indicate the DLX5+ GnRH neurons. (G) Schematic representation of a coronal section of a GW 8.5 fetal head. Squared box is magnified in (H) and it highlights the nasal region where simultaneous immunofluorescence for GnRH (green) and fluorescent in situ hybridization for DLX5 (red) was performed. (I) High-power confocal photomicrograph showing GnRH/DLX5 co-expressing migratory neurons (arrowheads). Scale bars: (I) 120 µm, (K) 20 µm. Abbreviations: OB, olfactory bulb; OE, olfactory epithelium, VNN/TN, vomeronasal nerve/terminal nerve; NS, nasal septum.
Wnt Inhibition Improves GnRH Neuron Differentiation Efficiency
From the pseudotime analysis, we also noticed that there is a cluster of genes downregulated along the GnRH lineage (Supplementary Fig. S8A). We performed an enrichment analysis and found that those genes were associated with sensory organ development, regulation of neurogenesis, and regulation of Wnt signaling pathway (Supplementary Fig. S9). Protein–protein interaction networks40 indicated that WNT5B has potential interactions with neuron progenitor marker genes and cell proliferation genes (Fig. 5A). These raised the intriguing possibility that Wnt inhibition might affect the yield of GnRH neurons. Therefore, we administrated XAV939, a Wnt inhibitor, during dSMADi stage (D0 to D10), and examined its effect at the end of differentiation (Fig. 5B). Interestingly, GNRH1 expression was upregulated significantly by XAV939 treatment. Concomitantly, the expressions of DLX1, DLX5, and GAD2 were increased in XAV939 treatment (Fig. 5C). These indicate that Wnt inhibition might enhance the function of GnRH neurons or improve the differentiation efficiency without affecting GnRH neuron properties. To investigate whether Wnt inhibition improves differentiation efficiency of GnRH neurons, we performed these 2 protocols on our GnRH reporter cell line, and used flow cytometry analysis to quantify the GnRH neurons. The results showed that the population of GnRH neurons (tdTomato+) increased over 3-fold in XAV939-treated cells (9.7 ± 0.7%) compared with dSMADi samples (37.8 ± 0.2%) (Fig. 5D). Consistently, the immunostaining of the differentiated cells also showed that there are more GnRH-positive neurons in XAV939-treated samples than dSMADi samples (Fig. 5E). Human GnRH is translated as a 96-amino-acid prohormone, which is processed by intracellular proteases to form the secreted decapeptide. We measured the secreted GnRH decapeptide of both conditions (XAV939 and dSMADi samples) in culture medium from D21 to D27. In line with our previous report,11 the secreted GnRH decapeptide was detectable since D21, but significantly increased from D25 onward. Moreover, XAV939-treated cells secreted more GnRH decapeptide than control samples (Fig. 5F), which also indicates that Wnt inhibition produced more functional GnRH neurons. To test whether the effect of Wnt inhibition is cell line-dependent, we repeated the differentiation in 2 additional hiPSC lines established from healthy donors (HEL11.441 and HEL24.342) and found that Wnt inhibition increased the GNRH1 expression in both hiPSC lines (Supplementary Fig. S10). In conclusion, Wnt inhibition during the dSMADi stage increases the yield and the secretion of GnRH neurons.
Figure 5.
Wnt inhibition improves the efficiency of GnRH neuron differentiation. (A) Protein–protein interaction analysis of downregulated genes in GnRHGABA branch. (B) Schematic of the differentiation protocols w/o XAV939 treatment. (C) RT-qPCR results show that DLX1, DLX5, and GAD2 are upregulated with Wnt inhibition along with GNRH1. The mRNA expression levels were compared with D0 samples. (D) Quantification of the ratio of generated GnRH neurons from flow cytometry analysis results. (E) Immunofluorescence staining shows that there are more GnRH neurons (green) generated with Wnt inhibition (XAV939). Arrows indicate GnRH neurons. Scale bar = 100 µm. (F) Secretion of GnRH decapeptides were measured with ELISA. D = days of differentiation. Data are represented as mean ± SD. Asterisks indicate the following: *P < .05; **P < .01; ***P < .001.
Wnt Inhibition Promotes Anterior Ventral GABAergic Neuron Specification
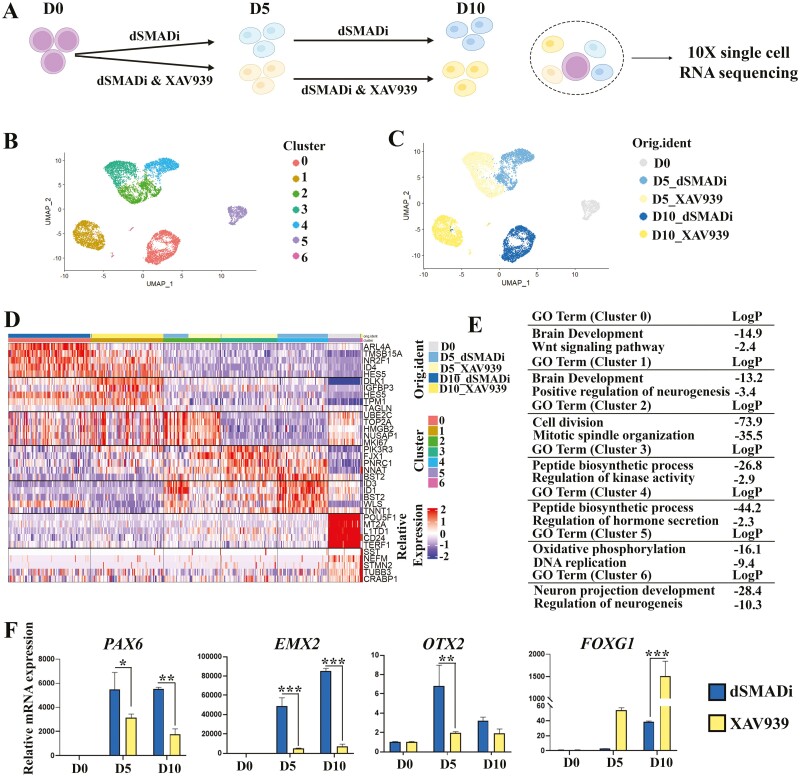
To investigate the mechanism that underlying Wnt inhibition improves differentiation efficiency of GnRH neurons, we performed scRNA-seq with the samples w/o XAV939 treatment (Fig. 6A), including undifferentiated sample (D0, 1008 cells analyzed), differentiating samples treated with only dSMADi (D5_dSMADi, 2480 cells analyzed, and D10_dSMADi, 2778 cells analyzed), and differentiating samples treated with both dSMADi and XAV939 (D5_XAV939, 2919 cells analyzed, and D10_XAV939, 2638 cells analyzed) (Fig. 6A). With Seurat analysis, we identified 7 cell clusters (Fig. 6B). Surprisingly, the samples are not very heterogeneous, and Wnt inhibition could already exhibit patterning function as early as D5 (Fig. 6B and 6C). From the heatmap showing the top 5 markers of each cell cluster (Fig. 6D, Supplementary Table S3), we noticed that cluster 0 is mainly from D10_dSMADi, cluster 1 mainly from D10_XAV939, and cluster 6 from both D10_XAV939 and D10_dSMADi. Cluster 3 is mainly from D5_XAV939, cluster 4 from D5_dSMADi, and cluster 2 from both D5_XAV939 and D5_dSMADi. Cluster 5 constituted mostly undifferentiated stem cells. By performing enrichment analysis, brain development is enriched in both cluster 0 and cluster 1, cell division is enriched in cluster 2, peptide biosynthetic process is enriched in both cluster 3 and cluster 4, oxidative phosphorylation process and DNA replication are enriched in cluster 5, and neuron projection development is enriched in cluster 6 (Fig. 6E). Based on the DEGs, XAV939 samples show higher FOXG1(anterior neural marker), DLK1, and PIK3R3 expression, while the dSMADi samples express not only higher Wnt signaling-related genes, including WLS, SFRP2, and WNT2B but also OTX2 (forebrain/midbrain neural marker), PAX6 (dorsal telencephalic progenitor marker), LHX5, and EMX2 (dorsal telencephalic marker) (Supplementary Fig. S11). Meanwhile, the expression of ventral neural markers, like HES1, ASCL1, GAD1/2, and DLX1/2, are slightly increased in XAV939 samples (Supplementary Fig. S11C). Through RT-qPCR, we validated that Wnt inhibition could increase the FOXG1 expression, and inhibit PAX6, OTX2, and EMX2 expressions (Fig. 6F). In addition to the patterning effect on stem cells, D10_XAV939 contributes a bigger cell portion to neural progenitors (Cluster 6, which is marked by higher expression of STMN2, NEUROG1, and NEUROD1) than D10_dSMADi (Fig. 6B-6D). In addition, Wnt inhibition samples had higher expression level of marker genes of NNPs, including DLK1 and FOXG1 than dSMADi samples on D20 (Supplementary Fig. S12). Thus, the administration of Wnt inhibition could promote anterior ventral GABAergic neuron fate specification and neuron differentiation progress in our cell model.
Figure 6.
Wnt inhibition promotes anterior ventral neuron specification and neurogenesis progress. (A) Schematic of the sample collection for scRNA-seq. (B) UMAP projections of cells from all 5 samples. Cells are colored according to their assigned cluster. (C) UMAP projections of cells from all 5 samples. Cells are colored according to sample names. (D) Heatmap of genes significantly enriched within each individual cluster. Single cells are shown in columns; genes are shown in rows. Top 5 differential genes of each cell cluster are shown. Bars for the cell clusters and sample names are both provided. (E) Representative enriched GO terms corresponding to the DEGs of each cluster. (F) RT-qPCR results shows that dorsal telencephalic progenitor markers PAX6, OTX2, and EMX2 are downregulated, and ventral telencephalic progenitor marker FOXG1 is upregulated with Wnt inhibition. The mRNA expression levels were compared with D0 samples. D = days of differentiation. Asterisks indicate the following: *P < .05; **P < .01; ***P < .001.
Discussion
GnRH neurons originate from the medial part of the nasal placode, vomeronasal organ (VNO), which in human fetuses contains actively proliferating (MKI67+/SOX2+) and nonproliferating progenitors (MKI67-/SOX2+).3 We identified 2 NP clusters in our in vitro model, consistent with the previous findings in vivo.3 Both NP clusters (NNPs and PNPs) expressed SOX2, NEUROG1, and NEUROD1 which are marker genes of neuronal progenitors in rodents.26-29 This suggests that the NPs generated in vitro show similar gene expression patterns to the NPs in vivo. Our data also suggest that NNPs are probably post-mitotic neural progenitors, which derived from PNPs. Based on their transcriptional pattern, different cell fates have been assigned to NNPs as a consequence of the fate-priming effect that typically occurs at the cell cycle exit or shortly thereafter.43 In this stage, selector and terminal differentiation genes activate differentiation cassettes that are sets of genes guiding the neuron to exhibit its fate-specific function.43 For instance, our data show that one sub-cluster of NNPs (NNP-C2) highly expresses GNRH1, and the expression of DLXs could possibly mark GnRHGABA precursors. Along the decision-making pathway, the cells will mainly differentiate into excitatory Glut neurons and to a lesser degree into inhibitory GnRHGABA neurons. Parallel to our finding, mouse GnRH neurons are also characterized as GABAergic neurons,44-46 which suggests conservation across mammalian species. From the scRNA-seq, we also observed a small cell cluster, which specifically expresses IRX family genes. This cluster can be an intermediate state between PNPs and NNPs based on trajectory analysis. In line with this, Irx has been reported to contribute to neural patterning.47 NPs are supposed to have the capacity to differentiate into both neuronal and glial cell types; however, we did not observe any oligodendrocytes or astrocytes. It is probably because we aimed to generate neurons in the end, and have not exposed the NPs with gliogenic fate stimulators such as neurotrophic factors (CNTF) or proteins (BMP, EGF, and FGF2)48,49 during differentiation.
An important finding of this study was the identification of the Glut cluster produced by this protocol. Besides characterizing the cell properties, we observed that the Glut cluster highly expresses microtubule protein Doublecortin (DCX), neural adhesion molecules (eg, NCAM1), and molecules helping neural migration (eg, RELN). Meanwhile, a previous study50 showed that GABA could slow GnRH neuron migration by depolarization via changes in intracellular chloride concentrations. Thus, GnRH neuron migration might occur through synergistic autocrine and paracrine signaling. Since Glut neurons are generated together with GnRHGABA, we hypothesized that they originated from the same or adjacent regions in humans. GnRH neurons are generated at the olfactory placode,4 a region that also gives rise to glutamatergic olfactory sensory neurons.51,52 According to the published data for human olfactory neurogenesis,38 olfactory sensory neurons are derived from NEUROG1 positive progenitors, and highly express GNG8, TUBB, EMX2, and LHX2 in immature states, which is consistent with the properties of the Glut cluster in our study. Several studies also suggested that Lhx2 is associated with olfactory receptor (OR) expression and is essential for OSNs identity53,54. However, we could not detect OR gene expression in our data (data not shown), which may be because of their immature state, as shown in human olfactory neurogenesis data,38 or insufficient sequencing depth. Further studies will be required to test whether Glut mimics the properties of olfactory sensory neurons in vivo.
From the integrated dataset, we identified 443 GnRH branch-associated genes (Supplementatry Fig. S8C, Table S2), including transcription factors, such as ISL1, DLX1/2, DLX5, FOXG1, SIX3, and ARX. Interestingly, they all associate with the olfactory epithelium or are involved in GnRH neuron development,8,13,55-62 which supports the reliability of our scRNA-seq analysis data. For example, ISL1 was found to be expressed in human fetal GnRH neurons and its expression was shown to persist during their migration to the hypothalamus.13 Isl1 expression was also observed in GnRH-3 neurons of terminal nerves in fish56 and in GnRH neurons in mice,54 but its ultimate function remains obscure as conditional ablation of Isl1 alone does not significantly compromise GnRH neuron migration or GnRH expression. This suggests that there are other compensatory mechanisms for the role of Isl1 in GnRH neuron development or another population of olfactory derived GnRH neurons, which are Isl1 negative.63 In this study, consistently with previous investigations in mice,58,59 we confirmed DLX5 expression in human GnRH neurons as well as in olfactory placode region. However, the role of Dlx5 in GnRH neuron development is not completely understood. Dlx5 seems to contribute to the activation of GnRH transcription directly in mouse GnRH neurons59 but also be required in building the connection of olfactory receptor neurons to the forebrain as in Dlx5−/− mice. GnRH neurons were present but failed to migrate into forebrain region in Dlx5−/− mice.58 Furthermore, Garaffo et al found that Dlx5 could promote the expression of miR-9 and miR-200 class, and depletion of miR-9 and mi-200 class in zebrafish results in both delayed differentiation of olfactory receptor neurons and impaired development of GnRH neurons.60 However, they could not find direct evidence of the function of Dlx5 in the genesis of GnRH neurons. Thus, further investigation on the role of DLX5 in human GnRH neuron development is needed.
We observed that modification of our original protocol with early Wnt inhibition significantly improved GnRH neuron differentiation efficiency. The increase of GNRH1 expression level (approximately 23-fold) and GnRH secretion at D25 (approximately 30-fold) were considerably higher than the increase of GnRH neuron yield (approximately 3-fold) in Wnt inhibition condition compared to plain dSMADi condition. Thus, we hypothesize that the GnRH neurons generated with Wnt inhibition are able to express higher levels of GNRH1 than the ones generated with dSMADi. By investigating the underlying mechanisms, we found that Wnt inhibition during dSMADi stage promotes neurogenesis, which is in agreement with previous studies showing that Wnt signaling enhances self-renewal of neural progenitors64 and that inhibition of canonical Wnt signaling is required for neuronal differentiation.65 Additionally, Wnt-inhibited samples express higher FOXG1 and SIX3, but lower PAX6, OTX2, and EMX2. These are all neural progenitor markers for different regional nervous system identity. In line with our findings, previous studies show that Wnt signaling specifies neural progenitor regional identity17 and that Wnt inhibition can induce a more ventralized gene expression profile including the augmentation of DLX2, DLX5, and FOXG1 expression.66,67 It has been reported that DLX transcription factors are master regulators of the developing vertebrate brain, driving GABAergic neuron differentiation.68 We identified only one GABAergic neuron cluster in our scRNA-seq data, ie, GnRHGABA neurons. However, it is not known whether there are other types of telencephalic GABAergic neurons promoted by XAV939. For this, we checked the expression of cortical interneuron markers at the end of differentiation,69,70 including LHX8, HMGB1, BASP1, SST, and PVALB, in addition to the spiny neuron markers,71,72 such as PPP1R1B, FOXP1, BCL11B, CHRM4, PLXND1 (Supplementary Fig. S14). LHX8 was increased in XAV939 condition, and when considering its potential role in GnRH neuron differentiation,13 the upregulation could also be caused by the higher yield of GnRH neurons. Since most of the cortical interneuron and spiny neuron markers were expressed at low levels and had no significant difference between dSMADi and XAV939 samples, it appears that there are no significant number of cortical neurons or spiny neurons generated following XAV939 condition. A further systematic study with higher resolution would be beneficial to investigate all the generated cell types with the developed protocol.
In previous studies, Wnt signaling activity has been detected in frontonasal region in transgenic mice16 and sFRP1 (a modulator of Wnt signaling) was detected in the nasal placode.15This suggests that Wnt signaling plays a role in olfactory development. Indeed, Wnt signaling has been demonstrated to be essential for promoting olfactory sensory neuronal fate choice.73 Thus, we suggest that during the embryonic olfactory neurogenesis, Wnt signaling could also play a role in patterning olfactory/GnRH neural fates. However, a recent study showed that Lgr4 deficiency results in a reduced number of GnRH neurons and subsequently delayed puberty in mice, presumably through impaired Wnt/β-catenin signaling.74 Another recent study shows that in human hypothalamus development, oligodendrocytes could interact with hypothalamic neurons (GnRH neurons were not included) to mediate neuronal maturation by Wnt, Hippo, and integrin signaling.75 These studies did not reveal the direct role of Wnt signaling in GnRH neuron development and more studies are warranted to its timely actions on GnRH neuron specification. However, we observed that Wnt inhibition during dSMADi promotes the differentiation of NPs and GABAergic neural fate specification, thus leading to a higher number of GnRH neurons. This finding provides a considerable advantage for further in vitro studies, and is potentially clinically important given that endocrine disruptors such as bisphenol A are reported to inhibit Wnt/β-catenin pathway in neurons76 and are implicated in central precocious puberty of girls.77
In conclusion, our work identifies a differentiation trajectory of human GnRH neurons in vitro and shows how another major neuron type (Glut) is generated together with GnRH neurons. We found that DLX family genes are specifically involved in the ontogeny of GnRH neurons, and demonstrate a robust optimization to the efficiency of this protocol, based on inhibition of Wnt signaling, which not only promotes NP differentiation but also specifies anterior ventral neuron fate. These findings delineate the molecular basis of human GnRH neuron development in vitro.
Supplementary Material
Acknowledgments
We would like to thank FIMM Single-Cell Analytics and Sequencing units supported by HiLIFE and Biocenter Finland for Single-Cell Sequencing services; thank Biomedicum FACS core, Biomedicum Imaging Unit, and Professor Timo Otonkoski at BSCC at the University of Helsinki, Finland, for providing resources, services, and cell lines; and thank Annika Tarkkanen for administration and language polishing. We thank the midwives of the Gynecology Department, Jeanne de Flandre Hospital of Lille (Centre d’Orthogenie), France, for their kind assistance and support; M Tardivel and A Bongiovanni (BICeL core facility of Lille, Univ. Lille, CNRS, Inserm, CHU Lille, Institut Pasteur de Lille, US 41-UMS 2014-PLBS, F-59000 Lille, France) for expert technical assistance. We acknowledge support of the Inserm Cross-Cutting Scientific Program (HuDeCA).
Contributor Information
Yafei Wang, Stem Cells and Metabolism Research Program, Research Programs Unit, and Department of Physiology, Faculty of Medicine, University of Helsinki, Helsinki, Finland.
Shrinidhi Madhusudan, Stem Cells and Metabolism Research Program, Research Programs Unit, and Department of Physiology, Faculty of Medicine, University of Helsinki, Helsinki, Finland.
Ludovica Cotellessa, Univ. Lille, Inserm, CHU Lille, Laboratory of Development and Plasticity of the Postnatal Brain, Lille Neuroscience & Cognition, UMR-S1172, Lille, France.
Jouni Kvist, Stem Cells and Metabolism Research Program, Research Programs Unit, and Department of Physiology, Faculty of Medicine, University of Helsinki, Helsinki, Finland.
Nazli Eskici, Stem Cells and Metabolism Research Program, Research Programs Unit, and Department of Physiology, Faculty of Medicine, University of Helsinki, Helsinki, Finland.
Venkatram Yellapragada, Stem Cells and Metabolism Research Program, Research Programs Unit, and Department of Physiology, Faculty of Medicine, University of Helsinki, Helsinki, Finland.
Kristiina Pulli, Stem Cells and Metabolism Research Program, Research Programs Unit, and Department of Physiology, Faculty of Medicine, University of Helsinki, Helsinki, Finland.
Carina Lund, Folkhälsan Research Center, Helsinki, Finland.
Kirsi Vaaralahti, Stem Cells and Metabolism Research Program, Research Programs Unit, and Department of Physiology, Faculty of Medicine, University of Helsinki, Helsinki, Finland; New Children’s Hospital, Pediatric Research Center, Helsinki University Hospital, Helsinki, Finland.
Timo Tuuri, Department of Obstetrics and Gynecology, Helsinki University Hospital, Helsinki, Finland.
Paolo Giacobini, Univ. Lille, Inserm, CHU Lille, Laboratory of Development and Plasticity of the Postnatal Brain, Lille Neuroscience & Cognition, UMR-S1172, Lille, France.
Taneli Raivio, Stem Cells and Metabolism Research Program, Research Programs Unit, and Department of Physiology, Faculty of Medicine, University of Helsinki, Helsinki, Finland; New Children’s Hospital, Pediatric Research Center, Helsinki University Hospital, Helsinki, Finland.
Funding
This work has been funded by Academy of Finland (315616 and 275259 to T.R.); Sigrid Juselius Foundation (T.R.); Novo Nordisk Fonden (NNF17OC0027448 to T.R.); Foundation for Pediatric Research (T.R.); The Hospital District of Helsinki and Uusimaa/Children and Adolescents (T.R.); European Union’s Horizon 2020 research and innovation program under the HORIZON EUROPE Marie Sklodowska-Curie Actions grant agreement (894596 to Y.W.); European Union’s Horizon 2020 research and innovation program under the HORIZON EUROPE Marie Sklodowska-Curie Actions grant agreement (813707 to S.M.); Paivikki ja Sakari Sohlberg Säätio (V.Y.); Instrumentarium Tiedesäätio (V.Y. and N.E). This work was supported by the Inserm Cross-Cutting Scientific Program (HuDeCA to P.G.), by the Agence Nationale de la Recherche (ANR), France (grant number ANR-19-CE16-0021-02 to P.G.).
Conflict of Interest
The authors indicated no financial relationships.
Author Contributions
Y.W.: conception and design, provision of study material, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. S.M., L.C., N.E., C.L.: provision of study material, collection and/or assembly of data, data analysis and interpretation, manuscript writing. J.K.: data analysis and interpretation. V.Y., K.P., K.V., T.T.: conception and design, manuscript writing. P.G.: financial support, administrative support, provision of study material, final approval of manuscript. T.R.: conception and design, financial support, administrative support, provision of study material, manuscript writing, final approval of manuscript.
Data Availability
All data needed to evaluate the conclusions in the paper are present in the paper and the Supplementary Materials. The sequencing data have been deposited in the Gene Expression Omnibus under accession number GSE212901.
References
- 1. Schwanzel-Fukuda M, Pfaff DW.. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338(6211):161-164. 10.1038/338161a0 [DOI] [PubMed] [Google Scholar]
- 2. Wray S, Grant P, Gainer H.. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci USA. 1989;86(20):8132-8136. 10.1073/pnas.86.20.8132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Casoni F, Malone SA, Belle M, et al. Development of the neurons controlling fertility in humans: new insights from 3D imaging and transparent fetal brains. Development. 2016;143(21):3969-3981. 10.1242/dev.139444 [DOI] [PubMed] [Google Scholar]
- 4. Cho HJ, Shan Y, Whittington NC, Wray S.. Nasal placode development, GnRH neuronal migration and Kallmann Syndrome. Front Cell Dev Biol. 2019;7:121. 10.3389/fcell.2019.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marques P, Skorupskaite K, Rozario KS, et al. . Physiology of GnRH and Gonadotropin secretion. Endotext. 2000. [Google Scholar]
- 6. Cangiano B, Swee DS, Quinton R, Bonomi M.. Genetics of congenital hypogonadotropic hypogonadism: peculiarities and phenotype of an oligogenic disease. Hum Genet. 2021;140(1):77-111. 10.1007/s00439-020-02147-1 [DOI] [PubMed] [Google Scholar]
- 7. Lee VH, Lee LT, Chow BK.. Gonadotropin-releasing hormone: regulation of the GnRH gene. FEBS J. 2008;275:5458-5478. 10.1111/j.1742-4658.2008.06676.x [DOI] [PubMed] [Google Scholar]
- 8. Messina A, Langlet F, Chachlaki K, et al. A microRNA switch regulates the rise in hypothalamic GnRH production before puberty. Nat Neurosci. 2016;19(6):835-844. 10.1038/nn.4298 [DOI] [PubMed] [Google Scholar]
- 9. Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861-872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 10. Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145-1147. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- 11. Lund C, Pulli K, Yellapragada V, et al. Development of gonadotropin-releasing hormone-secreting neurons from human pluripotent stem cells. Stem Cell Rep. 2016;7(2):149-157. 10.1016/j.stemcr.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yellapragada V, Liu X, Lund C, et al. MKRN3 interacts with several proteins implicated in puberty timing but does not influence GNRH1 expression. Front Endocrinol (Lausanne). 2019;10:48. 10.3389/fendo.2019.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lund C, Yellapragada V, Vuoristo S, et al. Characterization of the human GnRH neuron developmental transcriptome using a GNRH1-TdTomato reporter line in human pluripotent stem cells. Dis Model Mech. 2020;13(3):dmm040105. 10.1242/dmm.040105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang F, Barbacioru C, Wang Y, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377-382. 10.1038/nmeth.1315 [DOI] [PubMed] [Google Scholar]
- 15. Leimeister C, Bach A, Gessler M.. Developmental expression patterns of mouse sFRP genes encoding members of the secreted frizzled related protein family. Mech Dev. 1998;75(1-2):29-42. 10.1016/s0925-4773(98)00072-0 [DOI] [PubMed] [Google Scholar]
- 16. Maretto S, Cordenonsi M, Dupont S, et al. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci USA. 2003;100(6):3299-3304. 10.1073/pnas.0434590100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moya N, Cutts J, Gaasterland T, Willert K, Brafman DA.. Endogenous WNT signaling regulates hPSC-derived neural progenitor cell heterogeneity and specifies their regional identity. Stem Cell Rep. 2014;3(6):1015-1028. 10.1016/j.stemcr.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Satija R, Farrell JA, Gennert D, Schier AF, Regev A.. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495-502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888-1902 e1821. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Browaeys R, Saelens W, Saeys Y.. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2020;17(2):159-162. 10.1038/s41592-019-0667-5 [DOI] [PubMed] [Google Scholar]
- 21. Cao J, Spielmann M, Qiu X, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566(7745):496-502. 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381-386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aran D, Looney AP, Liu L, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163-172. 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanchate NK, Gacobini P, Lhuillier P, et al. . SEMA3A,a gene involved in axonal pathfinding, is mutated in patient with Kallmann Syndrome. PLoS Genet. 2012; 8(8): e1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shao X, Liao J, Lu X, et al. scCATCH: automatic annotation on cell types of clusters from single-cell RNA sequencing data. iScience. 2020;23(3):100882. 10.1016/j.isci.2020.100882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Graham V, Khudyakov J, Ellis P, Pevny L.. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39(5):749-765. 10.1016/s0896-6273(03)00497-5 [DOI] [PubMed] [Google Scholar]
- 27. Panaliappan TK, Wittmann W, Jidigam VK, et al. Sox2 is required for olfactory pit formation and olfactory neurogenesis through BMP restriction and Hes5 upregulation. Development. 2018;145(2): dev153791. 10.1242/dev.153791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shaker T, Dennis D, Kurrasch DM, Schuurmans C.. Neurog1 and Neurog2 coordinately regulate development of the olfactory system. Neural Dev. 2012;7:28. 10.1186/1749-8104-7-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Packard A, Giel-Moloney M, Leiter A, Schwob JE.. Progenitor cell capacity of NeuroD1-expressing globose basal cells in the mouse olfactory epithelium. J Comp Neurol. 2011;519(17):3580-3596. 10.1002/cne.22726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu H, Tomasiewicz H, Magnuson T, Rutishauser U.. The role of polysialic acid in migration of olfactory bulb interneuron precursors in the subventricular zone. Neuron. 1996;16(4):735-743. 10.1016/s0896-6273(00)80094-x [DOI] [PubMed] [Google Scholar]
- 31. Tang J, Landmesser L, Rutishauser U.. Polysialic acid influences specific pathfinding by avian motoneurons. Neuron. 1992;8(6):1031-1044. 10.1016/0896-6273(92)90125-w [DOI] [PubMed] [Google Scholar]
- 32. Hack I, Bancila M, Loulier K, Carroll P, Cremer H.. Reelin is a detachment signal in tangential chain-migration during postnatal neurogenesis. Nat Neurosci. 2002;5(10):939-945. 10.1038/nn923 [DOI] [PubMed] [Google Scholar]
- 33. Fujioka T, Kaneko N, Ajioka I, et al. . beta1 integrin signaling promotes neuronal migration along vascular scaffolds in the post-stroke brain. EBioMedicine. 2017;16:195-203. 10.1016/j.ebiom.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bot N, Schweizer C, Ben Halima S, Fraering PC.. Processing of the synaptic cell adhesion molecule neurexin-3beta by Alzheimer disease alpha- and gamma-secretases. J Biol Chem. 2011;286(4):2762-2773. 10.1074/jbc.M110.142521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gleeson JG, Lin PT, Flanagan LA, Walsh CA.. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23(2):257-271. 10.1016/s0896-6273(00)80778-3 [DOI] [PubMed] [Google Scholar]
- 36. Francis F, Koulakoff A, Boucher D, et al. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999;23(2):247-256. 10.1016/s0896-6273(00)80777-1 [DOI] [PubMed] [Google Scholar]
- 37. Wierman ME, Kiseljak-Vassiliades K, Tobet S.. Gonadotropin-releasing hormone (GnRH) neuron migration: initiation, maintenance and cessation as critical steps to ensure normal reproductive function. Front Neuroendocrinol. 2011;32(1):43-52. 10.1016/j.yfrne.2010.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Durante MA, Kurtenbach S, Sargi ZB, et al. Single-cell analysis of olfactory neurogenesis and differentiation in adult humans. Nat Neurosci. 2020;23(3):323-326. 10.1038/s41593-020-0587-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sullivan KA, Silverman AJ.. The ontogeny of gonadotropin-releasing hormone neurons in the chick. Neuroendocrinology. 1993;58(6):597-608. 10.1159/000126598 [DOI] [PubMed] [Google Scholar]
- 40. von Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31(1):258-261. 10.1093/nar/gkg034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mikkola M, Toivonen S, Tamminen K, et al. Lectin from Erythrina cristagalli supports undifferentiated growth and differentiation of human pluripotent stem cells. Stem Cells Dev. 2013;22(5):707-716. 10.1089/scd.2012.0365 [DOI] [PubMed] [Google Scholar]
- 42. Trokovic R, Weltner J, Otonkoski T.. Generation of iPSC line HEL24.3 from human neonatal foreskin fibroblasts. Stem Cell Res. 2015;15(1):266-268. 10.1016/j.scr.2015.05.012 [DOI] [PubMed] [Google Scholar]
- 43. Hobert O, Carrera I, Stefanakis N.. The molecular and gene regulatory signature of a neuron. Trends Neurosci. 2010;33(10):435-445. 10.1016/j.tins.2010.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ahnert-Hilger G, John M, Kistner U, Wiedenmann B, Jarry H.. Immortalized gonadotropin-releasing hormone neurons secrete gamma-aminobutyric acid-evidence for an autocrine regulation. Eur J Neurosci. 1998;10(3):1145-1152. 10.1046/j.1460-9568.1998.00129.x [DOI] [PubMed] [Google Scholar]
- 45. Heger S, Seney M, Bless E, et al. Overexpression of glutamic acid decarboxylase-67 (GAD-67) in gonadotropin-releasing hormone neurons disrupts migratory fate and female reproductive function in mice. Endocrinology. 2003;144(6):2566-2579. 10.1210/en.2002-221107 [DOI] [PubMed] [Google Scholar]
- 46. Vastagh C, Schwirtlich M, Kwakowsky A, et al. The spatiotemporal segregation of GAD forms defines distinct GABA signaling functions in the developing mouse olfactory system and provides novel insights into the origin and migration of GnRH neurons. Dev Neurobiol. 2015;75(3):249-270. 10.1002/dneu.22222 [DOI] [PubMed] [Google Scholar]
- 47. Rodriguez-Seguel E, Alarcon P, Gomez-Skarmeta JL.. The Xenopus Irx genes are essential for neural patterning and define the border between prethalamus and thalamus through mutual antagonism with the anterior repressors Fezf and Arx. Dev Biol. 2009;329(2):258-268. 10.1016/j.ydbio.2009.02.028 [DOI] [PubMed] [Google Scholar]
- 48. Song MR, Ghosh A.. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat Neurosci. 2004;7(3):229-235. 10.1038/nn1192 [DOI] [PubMed] [Google Scholar]
- 49. Chandrasekaran A, Avci HX, Leist M, Kobolak J, Dinnyes A.. Astrocyte differentiation of human pluripotent stem cells: new tools for neurological disorder research. Front Cell Neurosci. 2016;10:215. 10.3389/fncel.2016.00215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Casoni F, Hutchins BI, Donohue D, et al. SDF and GABA interact to regulate axophilic migration of GnRH neurons. J Cell Sci. 2012;125(Pt 21):5015-5025. 10.1242/jcs.101675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Murphy GJ, Glickfeld LL, Balsen Z, Isaacson JS.. Sensory neuron signaling to the brain: properties of transmitter release from olfactory nerve terminals. J Neurosci. 2004;24(12):3023-3030. 10.1523/JNEUROSCI.5745-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tabor R, Friedrich RW.. Pharmacological analysis of ionotropic glutamate receptor function in neuronal circuits of the zebrafish olfactory bulb. PLoS One. 2008;3(1):e1416. 10.1371/journal.pone.0001416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Plessy C, Pascarella G, Bertin N, et al. Promoter architecture of mouse olfactory receptor genes. Genome Res. 2012;22(3):486-497. 10.1101/gr.126201.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hussainy M, Korsching SI, Tresch A.. Pseudotime analysis reveals novel regulatory factors for multigenic onset and monogenic transition of odorant receptor expression. Sci Rep. 2002;12(1):16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Taroc EZM, Katreddi RR, Forni PE.. Identifying Isl1 genetic lineage in the developing olfactory system and in GnRH-1 neurons. Front Physiol. 2020;11:601923. 10.3389/fphys.2020.601923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Aguillon R, Batut J, Subramanian A, et al. Cell-type heterogeneity in the early zebrafish olfactory epithelium is generated from progenitors within preplacodal ectoderm. Elife. 2018;7: e32041. 10.7554/eLife.32041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang L, Zhang H, Hu G, et al. An early phase of embryonic Dlx5 expression defines the rostral boundary of the neural plate. J Neurosci. 1998;18(20):8322-8330. 10.1523/JNEUROSCI.18-20-08322.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Merlo GR, Mantero S, Zaghetto AA, et al. The role of Dlx homeogenes in early development of the olfactory pathway. J Mol Histol. 2007;38(6):612-623. 10.1007/s10735-007-9154-x [DOI] [PubMed] [Google Scholar]
- 59. Givens ML, Rave-Harel N, Goonewardena VD, et al. Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. J Biol Chem. 2005;280(19):19156-19165. 10.1074/jbc.M502004200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Garaffo G, Conte D, Provero P, et al. The Dlx5 and Foxg1 transcription factors, linked via miRNA-9 and -200, are required for the development of the olfactory and GnRH system. Mol Cell Neurosci. 2015;68:103-119. 10.1016/j.mcn.2015.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pandolfi EC, Hoffmann HM, Schoeller EL, Gorman MR, Mellon PL.. Haploinsufficiency of SIX3 abolishes male reproductive behavior through disrupted olfactory development, and impairs female fertility through disrupted GnRH neuron migration. Mol Neurobiol. 2018;55(11):8709-8727. 10.1007/s12035-018-1013-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Taroc EZM, Prasad A, Lin JM, Forni PE.. The terminal nerve plays a prominent role in GnRH-1 neuronal migration independent from proper olfactory and vomeronasal connections to the olfactory bulbs. Biol Open. 2017;6(10):1552-1568. 10.1242/bio.029074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shan Y, Saadi H, Wray S.. Heterogeneous origin of gonadotropin releasing hormone-1 neurons in mouse embryos detected by islet-1/2 expression. Front Cell Dev Biol. 2020;8:35. 10.3389/fcell.2020.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Seib DR, Corsini NS, Ellwanger K, et al. Loss of Dickkopf-1 restores neurogenesis in old age and counteracts cognitive decline. Cell Stem Cell. 2013;12(2):204-214. 10.1016/j.stem.2012.11.010 [DOI] [PubMed] [Google Scholar]
- 65. Aubert J, Dunstan H, Chambers I, Smith A.. Functional gene screening in embryonic stem cells implicates Wnt antagonism in neural differentiation. Nat Biotechnol. 2002;20(12):1240-1245. 10.1038/nbt763 [DOI] [PubMed] [Google Scholar]
- 66. Strano A, Tuck E, Stubbs VE, Livesey FJ.. Variable outcomes in neural differentiation of human PSCs arise from intrinsic differences in developmental signaling pathways. Cell Rep. 2020;31(10):107732. 10.1016/j.celrep.2020.107732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Verani R, Cappuccio P, Spinsanti R, et al. Expression of the Wnt inhibitor Dickkopf-1 is required for the induction of neural markers in mouse embryonic stem cells differentiating in response to retinoic acid. J Neurochem. 2007;100(1):242-250. 10.1111/j.1471-4159.2006.04207.x [DOI] [PubMed] [Google Scholar]
- 68. Lindtner S, Catta-Preta R, Tian H, et al. Genomic resolution of DLX-orchestrated transcriptional circuits driving development of forebrain GABAergic neurons. Cell Rep. 2019;28(8):2048-2063 e2048. 10.1016/j.celrep.2019.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Anderson KM, Collins MA, Chin R, et al. Transcriptional and imaging-genetic association of cortical interneurons, brain function, and schizophrenia risk. Nat Commun. 2020;11(1):2889. 10.1038/s41467-020-16710-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li Z, Wang D, Guo W, et al. Identification of cortical interneuron cell markers in mouse embryos based on machine learning analysis of single-cell transcriptomics. Front Neurosci. 2022;16:841145. 10.3389/fnins.2022.841145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD.. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28(3):622-632. 10.1523/JNEUROSCI.2986-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Precious SV, Kelly CM, Reddington AE, et al. FoxP1 marks medium spiny neurons from precursors to maturity and is required for their differentiation. Exp Neurol. 2016;282:9-18. 10.1016/j.expneurol.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fletcher RB, Das D, Gadye L, et al. Deconstructing olfactory stem cell trajectories at single-cell resolution. Cell Stem Cell. 2017;20(6):817-830 e818. 10.1016/j.stem.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mancini A, Howard SR, Marelli F, et al. LGR4 deficiency results in delayed puberty through impaired Wnt/beta-catenin signaling. JCI Insight. 2020;5(11): e133434. 10.1172/jci.insight.133434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhou X, Lu Y, Zhao F, et al. Deciphering the spatial-temporal transcriptional landscape of human hypothalamus development. Cell Stem Cell. 2022;29(2):328-343 e325. 10.1016/j.stem.2021.11.009 [DOI] [PubMed] [Google Scholar]
- 76. Tiwari SK, Agarwal S, Tripathi A, Chaturvedi RK.. Bisphenol—a mediated inhibition of hippocampal neurogenesis attenuated by curcumin via canonical Wnt pathway. Mol Neurobiol. 2016;53(5):3010-3029. 10.1007/s12035-015-9197-z [DOI] [PubMed] [Google Scholar]
- 77. Leonardi A, Penta L, Cofini M, et al. Pericardial effusion as a presenting symptom of Hashimoto thyroiditis: a case report. Int J Environ Res Public Health. 2017;14(12):1576. 10.3390/ijerph14121576 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and the Supplementary Materials. The sequencing data have been deposited in the Gene Expression Omnibus under accession number GSE212901.