Abstract
Organ-on-a-chip (OOC) is an emerging interdisciplinary technology that reconstitutes the structure, function, and physiology of human tissues as an alternative to conventional preclinical models for drug screening. Over the last decade, substantial progress has been made in mimicking tissue- and organ-level functions on chips through technical advances in biomaterials, stem cell engineering, microengineering, and microfluidic technologies. Structural and engineering constituents, as well as biological components, are critical factors to be considered to reconstitute the tissue function and microenvironment on chips. In this review, we highlight critical engineering technologies for reconstructing the tissue microarchitecture and dynamic spatiotemporal microenvironment in OOCs. We review the technological advances in the field of OOCs for a range of applications, including systemic analysis tools that can be integrated with OOCs, multiorgan-on-chips, and large-scale manufacturing. We then discuss the challenges and future directions for the development of advanced end-user-friendly OOC systems for a wide range of applications.
Keywords: Organ-on-a-chip, Tissue engineering, Microengineering
Introduction
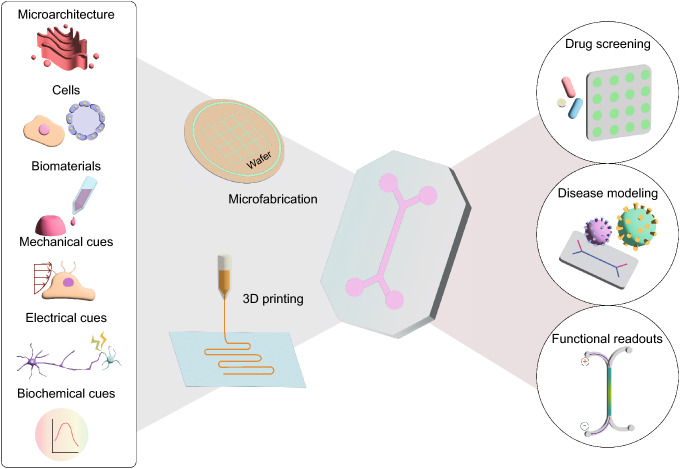
The high failure rates and long process of drug development are mainly attributed to the lack of human-relevant preclinical models with predictive power for clinical trials [1]. Advances in cell biology, microengineering, and tissue engineering have led to the emergence of an organ-on-a-chip (OOC) system that is designed to recapitulate key functional characteristics of living human organs [2] (Fig. 1). An OOC, which is also known as a microphysiological system, is a microengineered device that encompasses human-derived cells and physiologically relevant physicochemical microenvironments to reconstitute the microarchitecture and function of tissues [3]. OOC technology has offered the ability to accelerate clinical translational research by bridging the gap between animal studies and clinical trials.
Fig. 1.
From engineering considerations to applications of OOC. OOC integrates biological, biochemical, and engineering features of human organs in a microfluidic platform that is fabricated using cleanroom-based microfabrication or 3D printing technology. These OOCs can be used in a wide range of biomedical applications including drug screening, disease modeling, and monitoring tissue function
In the last decade, significant progress has been made in the development of OOC models to mimic a variety of human tissues (Table 1). In the context of tissue engineering, substantial efforts have been made to culture appropriate human-derived cell types in appropriate biomaterials for a stable and precise recreation of tissue-specific models. Advances in stem cell engineering, particularly in induced pluripotent stem cell (iPSC) technology, have further enabled the establishment of patient-specific OOC models for patient-specific preclinical studies [4]. Moreover, many types of biomaterials, including natural and synthetic biomaterials, have been investigated to reproduce the in vivo extracellular matrix (ECM) with tissue-specific properties [5].
Table 1.
Current progress in OOC research
| Tissue | Research objectives | Details of design | Future directions | Year | References |
|---|---|---|---|---|---|
| BBB | Measuring permeability and TEER with different concentrations of various molecules | A double layered microfluidic device with a sandwiched porous membrane |
- 3D culture of cells in a human brain-specific ECM - Co-culture of multiple human neurovascular unit (NVU) cells such as brain endothelial cells, pericytes, astrocytes, neurons, and microglia |
2012 | [125] |
| Quantification of 3D nanoparticle distribution in the vascular and perivascular regions at the BBB | A hybrid design combining two vertical microfluidic layers with three parallel microchannels aligned with microposts | 2020 | [9] | ||
| Developing an infectious human brain disease model to examine neurotropism and BBB penetration of pathogens |
- A vertical microfluidic chip with a gravity-driven unidirectional flow - Three parallel microchannels with small posts |
2021 | [126] | ||
| Heart | Recapitulating a physiological environment of native myocardium |
- Two compartmentalized microchambers separated by a PDMS membrane - Top compartment: subdivided by two rows of hanging posts for hosting cell construct - Bottom compartment: connected to a pneumatic actuation system to mimic beating motion |
- 3D culture of human stem cell-derived cardiac myocytes - Co-culture of myocytes with other cell types (fibroblasts and endothelial cells etc.) to recapitulate the multicellular heart |
2016 | [67] |
| Mimicking ischemia–reperfusion injury to evaluate cardioprotective effects of endothelial extracellular vesicles | 3D Printed multilayered PDMS films with an embedded flexible strain gauge | 2020 | [127] | ||
| Lung | Reconstituting the functional human alveolar-capillary interface |
- Two microchannels separated by a porous flexible membrane - Two lateral microchambers to apply vacuum that causes pressure-driven stretching of a membrane |
- Mimicking surfactant lining layer at the air–liquid interface - Using a biodegradable membrane that has tunable biochemical and mechanical properties |
2010 | [90] |
| Recapitulating the microstructure, ECM properties, air-cell interface, and the breathing motion the human pulmonary alveoli | A compartmentalized microchannels bonded with a 3D porous hydrogel showing an inverse opal structure | 2021 | [128] | ||
| Reproducing the biochemical and physical properties of the alveolar basal membrane with in vivo-like dimensions |
- An array of alveoli with in vivo-like dimensions - A stretchable biological membrane composed of proteins of the lung ECM |
2021 | [129] | ||
| Gut | Recapitulating the intestinal epithelium with the villi-like structure and the intestinal microbes |
- Two microchannels separated by a porous flexible membrane - Two lateral microchambers to apply vacuum that causes pressure-driven stretching of a membrane |
Adding other tissue-specific cell types such as mesenchymal cells or vascular cells | 2012 | [65] |
| Studying the mechanisms of the villi-like epithelial morphogenesis | 2019 | [10] | |||
| Liver | Constructing the heterogeneous lobule-mimetic pattern with liver cells | A concentric-stellate-tip electrode array with enhanced field-induced dielectrophoresis electrodes | Co-culture of key non-parenchymal cells | 2006 | [130] |
| Recapitulation of the structure and biological hierarchy of the liver tissue in a long-term cultured device | Hexagonal culture chamber that mimics the radial flow from the portal vein to the central vein | 2017 | [21] | ||
| Studying the pathogenesis of steatosis and monitoring hepatocyte functionality | An array of interconnected hexagonal microwells | 2019 | [98] |
However, not only the biological elements but also the design and engineering aspects, such as the microarchitecture of the device, physicochemical cues, and integration of biosensors, need to be considered to faithfully reconstruct human tissues in vitro. For example, microfluidic channels within a device should be suitably designed to create tissue-specific structures and microenvironments. Fabrication techniques should also be considered as fabrication methods affect the design and performance of OOCs [6]. In microfluidic OOC devices, environmental factors including mechanical, electrical, and chemical cues need to be controlled to mimic the physiological and pathological microenvironments of tissues that regulate tissue morphogenesis and function [7]. Moreover, the incorporation of sensors into OOCs allows for real-time on-chip analysis as well as monitoring of tissue functionality and microenvironments [8].
In this review, we describe the technical advances used to engineer OOCs, highlighting engineering technologies to reconstitute the microarchitecture and microenvironment of tissues. We then cover recent advanced technologies in the OOC field for their extension to integrative systemic analysis, systemic automation, and commercialization. Lastly, we briefly discuss the direction of the future work to expand the field of OOC for a broader range of applications.
Engineering three-dimensional (3D) microarchitectures of tissue microenvironment
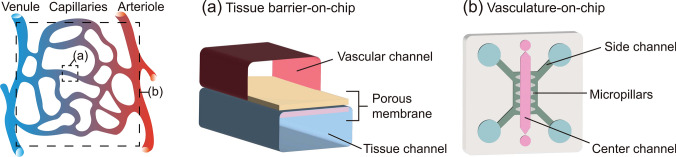
Microengineering technologies have enabled the precise in vitro reconstruction of tissue-specific microarchitectures. In particular, OOC devices are designed to recreate specific aspects of tissues for particular applications, such as studying drug transportation, monitoring barrier function, and studying cellular interactions in 3D. For drug screening, tissue barriers that control material transport between tissues were recreated to explore drug permeability and drug transport mechanisms. Double-layered microfluidic devices that have two parallel microfluidic layers compartmentalized by a porous membrane were used to recreate the barrier structures, including the blood–brain barrier (BBB) [9], intestinal mucosal barrier [10], and airway epithelial barrier [11] (Fig. 2a). The physically layered architecture of the device allows monitoring of the barrier integrity with transendothelial/epithelial electrical resistance (TEER) in real-time [9–11] and allows independent access to each layer for quantitative analysis of molecular distribution [9]. Moreover, fluid or air flow can be independently controlled in vertically separated microfluidic channels. A microengineered 3D vascular network-on-chip, which consists of multiple parallel channels laterally connected with arrays of micropillars, has been developed to recapitulate cell–cell interactions [12], vasculogenesis [13], and angiogenesis [14] in 3D (Fig. 2b). The array of micropillars between two parallel channels provides a semi-partitioned structure in which a liquid–solid interface can be formed by surface tension [15]. Taking advantage of the 3D microenvironment, this type of device has been used to study direct cell–cell and cell–material interactions under 3D co-culture conditions [16], particularly to investigate the tumor vascular niche [17, 18].
Fig. 2.
Two different types of OOCs mimicking vascular microenvironments. a Tissue barrier-on-a-chip consisting of two microfluidic layers with a porous membrane between the layers. b Vasculature-on-a-chip mimicking 3D vascular networks with parallel microchannels semi-partitioned by arrays of micropillars
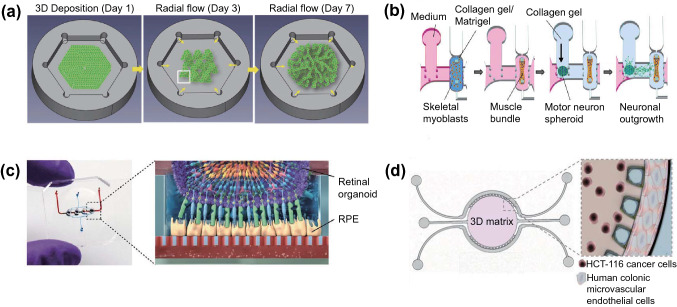
Different types of OOCs have been designed to reproduce tissue-specific microarchitectures. For example, human ocular surfaces including the cornea and conjunctiva were replicated in a multi-layered elastomeric device, where eye blinking was mimicked using a computer-controlled electromechanical actuator [19]. A field-induced dielectrophoresis-based cell patterning technique [20] and the hexagonal shape of a culture chamber with the concept of lattice growth [21] were used to mimic the structure of the liver lobules (Fig. 3a). Moreover, the highly organized anisotropic layers of the musculature of the heart were replicated by patterning microgrooves on soft multilayer cantilevers [22] and the neuromuscular junction platform was designed by co-culturing 3D skeletal muscle fiber bundles formed around the pillar structures and motor neuron spheroids embedded in collagen gel [23] (Fig. 3b).
Fig. 3.
Tissue-specific microarchitectures and biomaterials on chips. a Schematic illustration of liver-on-a-chip which mimics the structure of lattice of the liver lobules. Reproduced with permission from [21]. Copyright (2017) Wiley. b Schematic diagram of the 3D neuromuscular junction platform designed to co-culture 3D skeletal muscle fiber bundles and motor neuron spheroids. Reproduced with permission from [23]. Copyright (2018) AAAS. c Photo of the retina-on-a-chip (left) and schematic diagram of the retinal organoid and RPE in HA-based hydrogel on a chip (right). Reproduced with permission from [35]. Copyright (2019) eLife. d Schematic depiction and zoom-in image of the concept of colorectal tumor-on-a-chip. Reproduced with permission from [36]. Copyright (2019) AAAS
To fabricate the microarchitecture of OOC devices, cleanroom-based fabrication and 3D printing technology have been the most widely used among the various fabrication techniques [24–26]. Conventional cleanroom-based microfabrication, followed by soft lithography, allows the fabrication of high-resolution microstructures, whereas 3D printing technology enables the direct deposition of biological and functional materials, leading to a single-step fabrication of OOC devices [27–29]. Moreover, the integration of 3D bioprinting with microfluidics has enabled the creation of complex dynamic 3D microscale architectures in a high-throughput manner [30]. The evolution of 3D bioprinting technology has led to the development of printable bioink formulation [31]. Biomaterials presenting the required levels of rheological properties for 3D printing have been developed as 3D printable bioinks [32–34].
The selection of biomaterials with the desired physicochemical properties is important for mimicking the tissue-specific ECM [5, 37]. Physicochemical properties, such as surface chemistry, porosity, and stiffness profoundly affect the biochemical responses of cells that further regulate tissue homeostasis and pathogenesis [5]. Thus, various types of materials have been used to recreate tissue-specific ECMs to provide a physical and biological scaffold for cells [38]. Natural biomaterials, such as collagen, hyaluronic acid (HA), and Matrigel, are derived from living organisms and thus possess good biocompatibility. Collagen is the most prevalent protein in human tissues, particularly in connective tissues, including the bone, cartilage, skin, and cornea [39–41]. Owing to its insufficient mechanical strength, the extracted collagen is often combined with modified crosslinking methods or other high-strength materials to improve the physicochemical properties [39]. Collagen gel has been widely used to mimic the physiological environment of tumors in OOC devices, including a colorectal model [16] and a platform for studying breast ductal carcinoma [42]. HA is a glycosaminoglycan (GAG), which is an immunoneutral polysaccharide found throughout the human body [43]. HA hydrogels can be easily manipulated to possess biochemical functionality, dynamic microenvironments, and biological structures [44]. A human retina-on-a-chip model has incorporated HA-based hydrogels with retinal organoids and retinal pigment epithelia (RPE) to construct a retinal layer on a chip [35] (Fig. 3c). Matrigel is a solubilized basement membrane extracted from sarcomas and contains laminin, collagen, and heparan sulfate proteoglycan. Because of its components, Matrigel has been widely used to mimic the ECM of tumor [36] (Fig. 3d), stem cells [45], and neural tissues [9]. However, the undefined chemical composition and batch-to-batch variations of Matrigel make it difficult to reproducibly construct a tissue microenvironment [46]. Synthetic biomaterials are easily tunable to possess tissue-specific physicochemical properties [47, 48]. Moreover, synthetic biomaterials exhibit good reproducibility because they have less batch-to-batch variation than natural biomaterials [49]. Their lack of cell adhesion ligands can be supplemented by chemical modifications. Recently, hybrid natural–synthetic biomaterials that possess the advantages of both natural and synthetic biomaterials have been used as tissue scaffolds in OOCs [50].
More recently, decellularized ECM (dECM), a cell-removed remnant ECM of animal tissues or organs, has been used in OOCs to provide tissue-specific properties [51, 52]. In particular, dECM can restore the tissue-specialized spatial distribution of structural and functional ECM components. With the continuous development of decellularization protocols for various tissues, an increasing number of studies have introduced dECM into OOCs. For example, 3D liver dECM scaffolds were 3D printed to model hepatocellular carcinoma [53] and dECM from tumor-bearing and obese mammary glands were utilized for tumor cell studies [54].
In addition to physicochemical properties, electrical conductivity is an important feature of some tissues to be recapitulated in OOC. For electrically active tissue models, conductive biomaterials such as conductive hydrogels, conductive polymers, and carbon-based materials are used as scaffold materials [55]. Conductive hydrogels are representative materials for bioelectronic interfaces owing to their high stretchability, tissue-like softness, and high conductivity [56]. One study used a catechol-functionalized HA hydrogel with two electroconductive materials, single-walled carbon nanotubes (CNTs) and polypyrrole, as a conductive hydrogel for neuronal regeneration modeling [57]. Moreover, 3D printed CNTs based conductive scaffolds have been used for cardiac tissue engineering [58] and nerve regeneration modeling [59].
Dynamic control of tissue microenvironments
Cells in vivo are exposed to various physical stimuli, such as mechanical, electrical, and geometric actuations, as well as biochemical stimuli. In response to these stimuli, cells regulate their physiological and pathological functions. Hence, the need to recapitulate physiologically relevant extracellular stimuli has been underscored in developing in vitro tissue models [60]. In a microfluidics-based OOC model, physiological stimulation, such as fluidic shear stress, interstitial flow, cyclic strain of repetitive movements, and compression, can be precisely controlled and monitored in real-time [61] (Table 2).
Table 2.
Engineering technology to mimic specific features of tissue microenvironments
| Stimuli | Engineering technology | Mimicking tissue | Specific features | References |
|---|---|---|---|---|
| Shear stress | Integration of a microgap, self-contained flow loop, pneumatic pumps, and valves on a chip | Endothelial barrier | Pulsatile and oscillatory shear flow in pathogenic situations | [62] |
| Connection of the vascular channel to an external syringe pump | Endothelial barrier | Oscillatory shear stress caused by disturbed flow conditions in cardiovascular disease progression | [63] | |
| Cyclic strain | Stretching a porous membrane by applying vacuum to side channels | Lung | Cyclic breathing movement | [64] |
| Intestine | Intestinal peristaltic motions | [65] | ||
| Actuation of a micro-diaphragm using electro-pneumatic set-up | Lung | Cyclic breathing movements | [66] | |
| Pressurization of an auxiliary compartment that controls the membrane deformation | Cardiac tissue | Systolic and diastolic movements | [67] | |
| Electrical stimulus | Gold electrode patterning on PDMS microfluidic device | Cardiac tissue | Electrical pacing for function assessment | [73] |
| ITO-IDA electrode glass substrates on PDMS chip | Muscle | Tissue contraction during exercise | [74] | |
| 3D agar salt bridges and electrical circuit | Skin | Wound healing | [75] | |
| Biochemical gradients | Tree-like gradient generator | Neural tube | Early neural tube regionalization mediated by WNT signaling gradient | [76] |
| Placing the vasculature-mimicking lumen at one end of the chip | Solid tumor microenvironment | Acidic pH in tumor microenvironment | [78] | |
| Coating a gas impermeable film on the surface of the chip | Intestine | Steep oxygen gradient for healthy host-microbiome interactions | [79] |
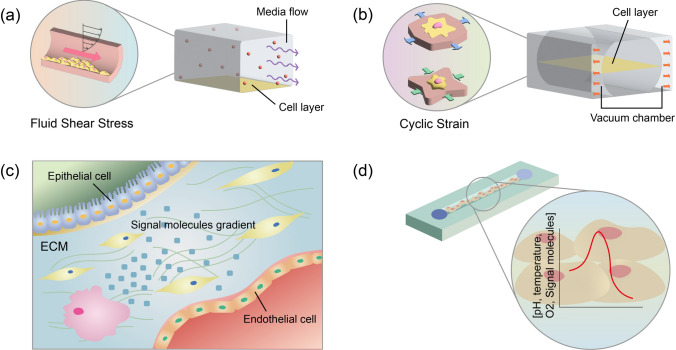
Flows in biological systems are replicated in a chip by flowing culture medium through microchannels with an auxiliary device such as a syringe pump and pneumatic pump [62]. The microscale channels of the OOC devices generate flows at low Reynolds numbers, resulting in laminar flow that can reproduce the general laminar and streamlined blood flow in the vasculature (Fig. 4a). Moreover, the flow of the culture medium along the microchannel helps maintain the long-term culture of the cells in the device by continuously refreshing the culture medium. Previous studies have shown that different flow conditions, including magnitude and oscillation frequency, affect the barrier integrity and inflammatory responses of endothelial cells (ECs), leading to pathogenic events [62, 63].
Fig. 4.
Mechanical and biochemical microenvironments of tissues. a, b Fluid shear stress a and cyclic strain b reproduced on a chip. c Biochemical microenvironment of tissues. (d) pH, temperature, and oxygen concentration gradients generated in OOC to mimic biochemical cues in tissue
Repetitive motions of organs, such as breathing motion of the lung and peristaltic motion in the gut, cause cyclic strain on the constituent cells. One representative OOC device mimicking cyclic strain motion utilized two side vacuum channels to stretch a porous membrane located in the center channel (Fig. 4b). This type of device was designed to construct various types of tissue models, including lung-on-a-chip [64] and gut-on-a-chip [65]. Another type of device which uses a micro-diaphragm that can be actuated by an electro-pneumatic setup was also developed to mimic 3D cyclic strain by breathing movements [66]. Moreover, the systolic and diastolic phases of the heart were replicated by pressurization of an auxiliary compartment that controls the membrane deformation between posts to induce homogeneous uniaxial cyclic strain [67].
Electrical stimulation is important for the development and regeneration of several tissues including neural [68], cardiac, and bone tissues [69]; wound healing [70, 71]; and cancer metastasis [72]. In the cardiac tissue chip model, gold electrodes were patterned and connected to the single generator MyoPacer for pacing cardiac tissue to assess cardiomyocyte function [73]. To construct an in vitro skeletal muscle tissue model of exercise training, a polydimethylsiloxane (PDMS) chip was bonded on glass substrates with indium tin oxide (ITO)-interdigitated array (IDA) electrodes that provide electrical stimulation [74]. An integrated microfluidic platform that can monitor real-time cell behavior under electrical stimuli and shear stress was designed to study the cellular migration of normal human dermal fibroblasts at the wound site [75]. In this device, 3D agar salt bridges were embedded in the platform at the end of the cell culture channel and connected to a single-loop electrical circuit to generate and control a constant electric field across the channel.
Microfluidic technologies enable the control of the spatiotemporal distribution of biochemical molecules to reconstitute various types of biochemical environments in tissues (Fig. 4c). A tree-like microfluidic gradient generator, one of the most widely used designs to form biochemical gradients, was integrated with a porous membrane-based double-layered microfluidic device for drug screening at different drug concentrations. Integration of the tree-like gradient generator has also enabled the modeling of neural tube development by providing a cranial rostral-to-caudal neural axis gradient [76]. Moreover, chemical gradients can be embedded into hydrogels to replicate the immobilized and soluble factor gradients incorporated in ECM [77].
Several other biochemical parameters, such as pH, oxygen, and temperature, control the physiological and pathological functions of cells and tissues (Fig. 4d). The pH acidification of the tumor microenvironment was recreated on a tumor-on-a-chip model by placing the vasculature-mimicking lumen at one end of the device to induce asymmetric distribution of nutrients and tumor cell viability [78]. In another study, a physiologically relevant hypoxic microenvironment was created by coating the surface of the OOC with a gas-impermeable film [79].
Functional readouts for integrated systemic analysis
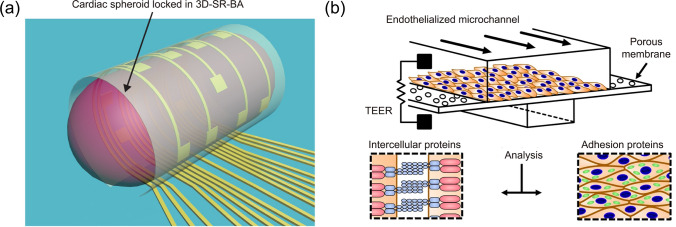
Integration of analytical biosensors into OOCs allows for real-time and on-chip monitoring of cells and their microenvironment; this is useful to measure the function of cells and tissues, analyze biological responses, or control the tissue microenvironment on the chip. Various types of physical and chemical sensors have been combined with OOCs to monitor the physiological and pathological conditions of tissues. For example, a muscular thin film (MTF) technique was integrated into a heart-on-a-chip to measure the contractility and action potential propagation of the anisotropic ventricular myocardium [80, 81]. Moreover, an organ-on-an-electronic-chip implementing 3D self-rolled biosensor arrays (3D-SR-BAs) was developed to record electrophysiological signals from human cardiac spheroids [82] (Fig. 5a). The liver-on-a-chip device was equipped with tissue-embedded oxygen sensors and connected to a sensor unit containing electrochemical sensors for real-time monitoring of the dynamics of mitochondrial dysfunction by measuring glucose and lactate metabolism [83].
Fig. 5.
Monitoring tissue function on a chip. a 3D-SR-BAs which monitors electrophysiological signal transduction of human cardiac spheroids. Reproduced with permission from [82]. Copyright (2019) AAAS. b TEER implemented chip that monitors barrier permeability on a chip in real-time. Reproduced with permission from [63]. Copyright (2017) Nature
The electrical activity of electrogenic tissues such as neuronal, muscle, and cardiac tissues can be recorded using microelectrode arrays (MEAs) [84–86]. In vitro MEAs contain more than 10,000 microelectrodes that record extracellular action potentials and stimulate cells at the single-cell level [87]. To minimize the mechanical mismatch at the interface between the cells and the electrodes, MEAs have been coated with bioactive hydrogels or fabricated with soft electrode materials [88]. Further, 3D printing technology has enabled the fabrication of high-resolution MEAs on soft substrate [88] and 3D microtower MEAs [85] in a time- and cost-efficient manner.
The barrier function of the tissue layer can be monitored using TEER in a real-time and non-invasive manner [89]. Many types of OOCs mimicking tissue barriers such as the BBB [9], lung epithelium [90], and intestinal barrier [65] have implemented TEER to monitor tissue barrier function over time. Previous studies measured TEER by repeatedly inserting electrodes into microfluidic channels [65, 90], which can cause large measurement variability. Further, the integration of electrodes within OOCs by coating the electrodes on the chip substrates has enhanced the stability and reliability of TEER measurements [63, 91] (Fig. 5b). One recent study showed a spatial-TEER-integrated OOC that can move along the length of the microchannel for localized measurements of TEER [92].
With the increasing demand for OOC platforms in the pharmaceutical field, several attempts have been made to integrate drug-screening tools with OOCs for on-chip analysis. An integrated microfluidic system consisting of a cell culture chamber, biochemical sensors, and fluid channels was developed to detect glucose and lactate production, pH, and oxygen levels in response to candidate drugs [93]. In one study, a microfluidic human serum albumin (HSA) immunosensor was embedded in a liver-on-a-chip to monitor albumin levels in diabetic disease modeling and drug treatment [94]. The cardiac muscle tissue chip integrated with MTF showed great potential for high-throughput drug-screening applications [95].
Advanced engineering technology in organs-on-chips
OOCs have been used to understand pathogenesis or pathophysiology of various diseases by mimicking pathological conditions of the diseases using iPSCs, disease-specific microarchitecture, or physicochemical microenvironments. For example, a 3D human Alzheimer's disease (AD) model that recapitulates the key features of AD (beta-amyloid aggregation, phosphorylated tau accumulation, and neuroinflammation) has been developed by culturing iPSC-derived AD neurons, astrocytes and microglia [96]. A 3D stenosis model with different constriction geometries that can recapitulate flow disturbances was developed to understand atherogenic flow-mediated endothelial dysfunction [97]. A non-alcoholic fatty liver disease (steatosis) has been modeled by culturing spheroids consisting of human hepatocellular carcinoma cells and human umbilical vein endothelial cells with free fatty acids supplementation [98].
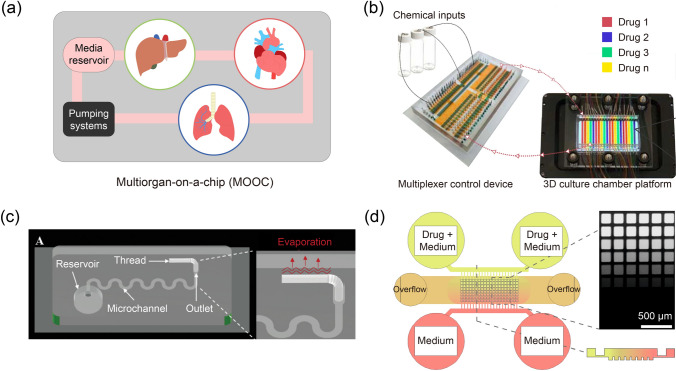
As multiple organs interact with each other to control many biological processes in the human body, multiorgan-on-a-chip (MOOC) systems that combine multiple OOCs have been developed [99, 100] (Fig. 6a). These models have great potential for modeling drug adsorption, distribution, metabolism, excretion, and toxicity (ADMET) on chips. Various types of MOOCs, such as liver-cancer-heart-on-a-chip [101], liver-heart–lung-on-a-chip [102], and liver-cardiac-skeletal muscle-neurons-on-a-chip [103] were developed to predict drug efficacy and toxicity. MOOC platforms can be used not only for drug discovery but also for studying a variety of multiorgan metabolic activities. A microfluidic platform that reconstructs the human female reproductive tract and peripheral tissues by integrating the ovary, uterus, fallopian tube, cervix, and liver was developed as a tool for studying hormonal signaling during the human menstrual cycle and pregnancy [104]. These MOOCs need to incorporate interconnecting flow system between the multiple organ microenvironments. A gastrointestine (GI)-liver-on-a-chip has implemented the oscillatory bidirectional fluidic flow system that utilize gravity to obtain a wide range of flow rates depending on the dimension of a microchannel [105]. Moreover, interconnection via vascularization and scaling of coupled OOCs are crucial factors to design MOOCs [106, 107].
Fig. 6.
Advanced engineering technology in OOCs. a Schematic illustration of liver-heart–lung-on-a-chip. b Automated microfluidic platform for high-throughput drug screening of tumor organoids. Reproduced with permission from [110]. Copyright (2020) Nature. c Pumpless and tubeless microfluidic chip that controls flow rates using adjustable hydrophilic thread. Reproduced with permission from [114]. Copyright (2020) Wiley. d A self-generated microfluidic concentration gradient chip for drug screening. Reproduced with permission from [115]. Copyright (2018) Nature
The increasing demand for OOCs has led to the need for the automation and commercialization of the system. Recently, a robotic microfluidic platform “interrogator” was developed to automatically perform cell culture, perfusion, medium addition, fluidic linking, sample collection, and imaging for MOOC that links up to ten vascularized OOCs [108]. Moreover, a robotic liquid transfer system was used to fluidically link multiple OOCs for pharmacokinetic (PK) and pharmacodynamic (PD) modeling [109]. Another example of an automated and high-throughput system was developed for the dynamic and real-time analysis of growth, morphological changes, cellular apoptosis, and death of organoids. This system consisted of a multiplexer control device, 3D culture chamber, and custom software to facilitate personalized drug screening [110] (Fig. 6b). For the technology to be commercialized, large-scale, high-throughput, and automated fabrication processes must be satisfied [111]. Injection molding has been widely used for the large-scale production of OOCs owing to its advantages including low cost and time efficiency. Once the device design and fabrication steps are optimized for injection modeling, the entire production process becomes considerably simpler and faster than conventional soft lithography. An injection-molded plastic array platform mimicking the 3D tumor lymphatic vascular network has shown great potential for high-throughput drug testing [112]. However, the range of materials that can be used for injection molding is limited [113].
Recent advances in microfluidic technologies have led to the development of user-friendly OOCs. For example, recent studies have suggested microfluidic devices that can introduce flow into microfluidic channels without using external instrumentation. A pumpless and tubeless OOC platform could obtain different flow rates by adjusting the length and diameter of the hydrophilic thread [114] (Fig. 6c). In a multilayered microfluidic device, the resistive and capacitive microfluidic network and overflow ports could create long-lasting self-generated microfluidic concentration gradient [115] (Fig. 6d). A long-lasting, self-generated drug concentration gradient could be formed across an array of 240 tumor spheroids in a device to perform drug screening [115].
Summary and perspectives
Recently, significant advances have been made in the field of OOC technologies, demonstrating great potential as a complementary tool to preclinical animal studies. Cell biology, in conjunction with the development of microengineering and tissue engineering, has contributed to rapid progress in OOC research. Design of the microscale devices and dynamic control of physicochemical cues in a device have provided tissue-specific microenvironments to precisely recapitulate the key features of tissues on chips. Moreover, integration of biosensors has allowed for monitoring of the biological responses of the tissue models.
Currently, OOC research, from cell culture to functional analysis, requires technical skills and training to handle with the microscale devices. One of the major challenges in the field is a high user dependency that causes low reproducibility of the device. With the increasing demand of OOCs in various fields, systemized experimental procedures need to be established to minimize the user dependency. Moreover, high-throughput and automated systems could open the way for manufacturing end user-friendly OOC systems for practical applications. It is also challenging to reconstitute the complex and dynamic 3D microenvironment of tissues which can be precisely controlled in real-time. Introducing smart biomaterials such as stimuli-responsive biomaterials [116] and 3D spatiotemporally manipulative biomaterials [117] into OOC devices may better reconstruct the dynamic 3D microenvironment that regulates a variety of events in tissues [117–119].
OOC platforms can be utilized in a wide range of applications as preclinical tools, mainly in drug screening and pathophysiological studies. In addition to the newly developed drugs, nanoparticle-based drug delivery systems can be evaluated for their toxicity [120] and targeting efficacy [121] using OOCs [122]. Recently, iPSC technology has opened a new door to establishing personalized OOC models to develop personalized medicine [4]. Moreover, OOC devices can be used to study the pathophysiology of highly infectious illnesses such as SARS-CoV-2 [123] and influenza virus infection [124]. In this regard, the OOC technology may provide a breakthrough in fighting future pandemic respiratory viruses. Another potential application is the preclinical testing of implantable devices. The convergence of microsensors and bioelectronics into OOCs can mimic the tissue-implant interface, permitting better evaluation of the safety and efficacy of devices before implantation.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2022R1C1C1010823) and Pusan National University Research Grant, 2021.
Declaration
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ma C, et al. Organ-on-a-chip: a new paradigm for drug development. Trends Pharmacol Sci. 2021 doi: 10.1016/j.tips.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huh D, Ingber GA, Donald E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011 doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park J, et al. Organomimetic microsystems technologies. Biomed Eng Lett. 2012 doi: 10.1007/s13534-012-0059-6. [DOI] [Google Scholar]
- 4.Vatine GD, et al. Human iPSC-derived blood-brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell. 2019 doi: 10.1016/j.stem.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 5.Osório LA, et al. A review of biomaterials and scaffold fabrication for organ-on-a-Chip (OOAC) Sys. Bioeng. 2021. 10.3390/bioengineering8080113 [DOI] [PMC free article] [PubMed]
- 6.Leung CM, et al. A guide to the organ-on-a-chip. Nat Rev Methods Primers. 2022 doi: 10.1038/s43586-022-00118-6. [DOI] [Google Scholar]
- 7.Young EW, Beebe DJ. Fundamentals of microfluidic cell culture in controlled microenvironments. Chem Soc Rev. 2010 doi: 10.1039/b909900j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B, Korolij A, Benjamin FLL, Radisic M. Advances in organ-on-a-chip engineering. Nat Rev Mat. 2018 doi: 10.1038/s41578-018-0034-7. [DOI] [Google Scholar]
- 9.Ahn SI, et al. Microengineered human blood-brain barrier platform for understanding nanoparticle transport mechanisms. Nat Commun. 2020 doi: 10.1038/s41467-019-13896-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin W, Ingber CD, Kim DE, Jung H. Human intestinal morphogenesis controlled by transepithelial morphogen gradient and flow-dependent physical cues in a microengineered gut-on-a-chip. iScience. 2019 doi: 10.1016/j.isci.2019.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Si L, et al. A human-airway-on-a-chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nat Biomed Eng. 2021 doi: 10.1038/s41551-021-00718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bang S, et al. A low permeability microfluidic blood-brain barrier platform with direct contact between perfusable vascular network and astrocytes. Sci Rep. 2017 doi: 10.1038/s41598-017-07416-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C, et al. Label-free three-dimensional observations and quantitative characterisation of on-chip vasculogenesis using optical diffraction tomography. Lab Chip. 2021 doi: 10.1039/D0LC01061H. [DOI] [PubMed] [Google Scholar]
- 14.Ahn Jungho, Sei Yoshitaka J., Jeon Noo Li, Kim YongTae. Probing the effect of bioinspired nanomaterials on angiogenic sprouting with a microengineered vascular system. IEEE Trans Nanotechnol. 2018 doi: 10.1109/TNANO.2017.2771426. [DOI] [Google Scholar]
- 15.Huang CP, et al. Engineering microscale cellular niches for three-dimensional multicellular co-cultures. Lab Chip. 2009 doi: 10.1039/b818401a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong S-Y, et al. Co-culture of tumor spheroids and fibroblasts in a collagen matrix-incorporated microfluidic chip mimics reciprocal activation in solid tumor microenvironment. PLoS ONE. 2016 doi: 10.1371/journal.pone.0159013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S, Koo S, Yu D-J, Cho J, Lee H, Song H, Kim JM, Min S-Y, Jeon D-H, Li N. 3D microfluidic platform and tumor vascular mapping for evaluating anti-angiogenic RNAi-based nanomedicine. ACS Nano. 2021;1:1. doi: 10.1021/acsnano.0c05110. [DOI] [PubMed] [Google Scholar]
- 18.Haase K, Gillrie GS, Kamm MR, Roger D. Endothelial regulation of drug transport in a 3D vascularized tumor model. Adv Funct Mater. 2020;1:1. doi: 10.1002/adfm.202002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo J, Byun WY, Alisafaei F, Georgescu A, Yi YS, Massaro-Giordano M, Shenoy VB, Lee V, Bunya VY, Huh D. Multiscale reverse engineering of the human ocular surface. Nat Med. 2019;1:1. doi: 10.1038/s41591-019-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho CT, et al. Liver-cell patterning lab chip: mimicking the morphology of liver lobule tissue. Lab Chip. 2013 doi: 10.1039/c3lc50402f. [DOI] [PubMed] [Google Scholar]
- 21.Weng YS, Chang SF, Shih MC, Tseng SH, Lai CH. Scaffold-Free Liver-On-A-Chip with Multiscale Organotypic Cultures. Adv Mater. 2017;1:1. doi: 10.1002/adma.201701545. [DOI] [PubMed] [Google Scholar]
- 22.Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan H, Yadid M, Park SJ, Kotikian A, Nesmith AP, Campbell PH, Vlassak JJ. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat Mater. 2017 doi: 10.1038/nmat4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamm TOGMUD. Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons. Sci Adv. 2018 doi: 10.1126/sciadv.aat5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson BN, et al. 3D printed nervous system on a chip. Lab Chip. 2016 doi: 10.1039/C5LC01270H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharjee N, et al. Desktop-stereolithography 3D-printing of a poly (dimethylsiloxane)-based material with sylgard-184 properties. Adv Mater. 2018 doi: 10.1002/adma.201800001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olanrewaju A, et al. Capillary microfluidics in microchannels: from microfluidic networks to capillaric circuits. Lab Chip. 2018 doi: 10.1039/C8LC00458G. [DOI] [PubMed] [Google Scholar]
- 27.Knowlton S, et al. Towards single-step biofabrication of organs on a chip via 3D printing. Trends Biotechnol. 2016 doi: 10.1016/j.tibtech.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Yi HG, et al. 3D printing of organs-on-chips. Bioengineering (Basel) 2017 doi: 10.3390/bioengineering4010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, et al. Freeform 3D printing of soft matters: recent advances in technology for biomedical engineering. Biomed Eng Lett. 2020 doi: 10.1007/s13534-020-00171-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datta P, Dey M, Ataie Z, Unutmaz D, Ozbolat IT. 3D bioprinting for reconstituting the cancer microenvironment. NPJ Precis. Oncol. 2020;1:1. doi: 10.1038/s41698-020-0121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gopinathan J, Noh I. Recent trends in bioinks for 3D printing. Biomater Res. 2018 doi: 10.1186/s40824-018-0122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan FLC, et al. Dynamic bioinks to advance bioprinting. Adv Healthcare Mater. 2020 doi: 10.1002/adhm.201901798. [DOI] [PubMed] [Google Scholar]
- 33.Schwab A, et al. Printability and shape fidelity of bioinks in 3D bioprinting. Chem Rev. 2020 doi: 10.1021/acs.chemrev.0c00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unagolla JM, Jayasuriya AC, et al. Hydrogel-based 3D bioprinting: A comprehensive review on cell-laden hydrogels, bioink formulations, and future perspectives. Appl Mater Today. 2020 doi: 10.1016/j.apmt.2019.100479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Achberger K, Probst C, Haderspeck J, Bolz S, Rogal J, Chuchuy J, Nikolova M, Cora V, Antkowiak L, Haq W, Shen N. Merging organoid and organ-on-a-chip technology to generate complex multi-layer tissue models in a human retina-on-a-chip platform. Elife. 2019 doi: 10.7554/eLife.46188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carvalho MR, et al. Colorectal tumor-on-a-chip system: a 3D tool for precision onco-nanomedicine. Sci Adv. 2019 doi: 10.1126/sciadv.aaw1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikolova MP, Chavali MS. Recent advances in biomaterials for 3D scaffolds: a review. Bioactive Mater. 2019 doi: 10.1016/j.bioactmat.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hussey GS, Dziki JL, Badylak SF. Extracellular matrix-based materials for regenerative medicine. Nat Rev Mater. 2018 doi: 10.1038/s41578-018-0023-x. [DOI] [Google Scholar]
- 39.Lin K, Zhang D, Macedo MH, Cui W, Sarmento B, Shen G. Advanced collagen-based biomaterials for regenerative biomedicine. Adv Func Mater. 2019 doi: 10.1002/adfm.201804943. [DOI] [Google Scholar]
- 40.Cen L, Liu WEI, Cui LEI, Zhang W, Cao Y. Collagen tissue engineering: development of novel biomaterials and applications. Pediat Res. 2008 doi: 10.1203/PDR.0b013e31816c5bc3. [DOI] [PubMed] [Google Scholar]
- 41.Sorushanova A, Delgado LM, Wu Z, Shologu N, Kshirsagar A, Raghunath R, Mullen AM, Bayon Y, Pandit A, Raghunath M, Zeugolis DI. The collagen suprafamily: from biosynthesis to advanced biomaterial development. Adv Mater. 2019 doi: 10.1002/adma.201801651. [DOI] [PubMed] [Google Scholar]
- 42.Choi Y, et al. A microengineered pathophysiological model of early-stage breast cancer. Lab Chip. 2015 doi: 10.1039/C5LC00514K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burdick JA, Prestwich GD. Hyaluronic acid hydrogels for biomedical applications. Adv Mater. 2011 doi: 10.1002/adma.201003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Highley CB, et al. Recent advances in hyaluronic acid hydrogels for biomedical applications. Curr Opin Biotechnol. 2016 doi: 10.1016/j.copbio.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Gkatzis K, et al. Use of three-dimensional organoids and lung-on-a-chip methods to study lung development, regeneration and disease. Eur Respir J. 2018 doi: 10.1183/13993003.00876-2018. [DOI] [PubMed] [Google Scholar]
- 46.Kozlowski MT, et al. Towards organoid culture without Matrigel. Commun Biol. 2021 doi: 10.1038/s42003-021-02910-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aisenbrey EA, Murphy WL. Synthetic alternatives to Matrigel. Nat Rev Mater. 2020 doi: 10.1038/s41578-020-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xing H, Lee H, Luo L, Kyriakides TR. Extracellular matrix-derived biomaterials in engineering cell function. Biotechnol Adv. 2020 doi: 10.1016/j.biotechadv.2019.107421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahadian S, et al. Organ-on-a-chip platforms: a convergence of advanced materials, cells, and microscale technologies. Adv Healthc Mater. 2018 doi: 10.1002/adhm.201700506. [DOI] [PubMed] [Google Scholar]
- 50.Setayeshmehr M, et al. Hybrid and composite scaffolds based on extracellular matrices for cartilage tissue engineering. Tissue Eng Part B Rev. 2019 doi: 10.1089/ten.teb.2018.0245. [DOI] [PubMed] [Google Scholar]
- 51.Kim BS, Das S, Jang J, Cho DW. Decellularized extracellular matrix-based bioinks for engineering tissue- and organ-specific microenvironments. Chem Rev. 2020 doi: 10.1021/acs.chemrev.9b00808. [DOI] [PubMed] [Google Scholar]
- 52.Kim BS, Kwon YW, Kong JS, Park GT, Gao G, Han W, Kim MB, Lee H, Kim JH, Cho DW. 3D cell printing of in vitro stabilized skin model and in vivo pre-vascularized skin patch using tissue-specific extracellular matrix bioink: A step towards advanced skin tissue engineering. Biomaterials. 2018 doi: 10.1016/j.biomaterials.2018.03.040. [DOI] [PubMed] [Google Scholar]
- 53.Ma X, Yu C, Wang P, Xu W, Wan X, Lai CSE, Liu J, Koroleva-Maharajh A, Chen S. Rapid 3D bioprinting of decellularized extracellular matrix with regionally varied mechanical properties and biomimetic microarchitecture. Biomaterials. 2018 doi: 10.1016/j.biomaterials.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wishart AL, Conner SJ, Guarin JR, Fatherree JP, Peng Y, McGinn RA, Crews R, Naber SP, Hunter M, Greenberg AS, Oudin MJ. Decellularized extracellular matrix scaffolds identify full-length collagen VI as a driver of breast cancer cell invasion in obesity and metastasis. Sci Adv. 2020 doi: 10.1126/sciadv.abc3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou J, Vijayavenkataraman S. D-printable conductive materials for tissue engineering and biomedical applications. Bioprinting. 2021 doi: 10.1016/j.bprint.2021.e00166. [DOI] [Google Scholar]
- 56.Yuk H, et al. Hydrogel bioelectronics. Chem Soc Rev. 2019 doi: 10.1039/c8cs00595h. [DOI] [PubMed] [Google Scholar]
- 57.Shin J, et al. Three-dimensional electroconductive hyaluronic acid hydrogels incorporated with carbon nanotubes and polypyrrole by catechol-mediated dispersion enhance neurogenesis of human neural stem cells. Biomacromol. 2017 doi: 10.1021/acs.biomac.7b00568. [DOI] [PubMed] [Google Scholar]
- 58.Ho CMB, Mishra A, Lin PTP, Ng SH, Yeong WY, Kim YJ, Yoon YJ. 3D printed polycaprolactone carbon nanotube composite scaffolds for cardiac tissue engineering. Macromol Biosci. 2017 doi: 10.1002/mabi.201600250. [DOI] [PubMed] [Google Scholar]
- 59.Lee SJ, Zhu W, Nowicki M, Lee G, Heo DN, Kim J, Zuo YY, Zhang LG. 3D printing nano conductive multi-walled carbon nanotube scaffolds for nerve regeneration. J Neural Eng. 2018 doi: 10.1088/1741-2552/aa95a5. [DOI] [PubMed] [Google Scholar]
- 60.Zuppinger C. 3D cardiac cell culture: a critical review of current technologies and applications. Front Cardiovasc Med. 2019 doi: 10.3389/fcvm.2019.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ergir E, Bachmann B, Redl H, Forte G, Ertl P. Small force big impact: next generation organ-on-a-chip systems incorporating biomechanical cues. Front Physiol. 2018 doi: 10.3389/fphys.2018.01417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shao J, Wu L, Wu J, Zheng Y, Zhao H, Jin Q, Zhao J. Integrated microfluidic chip for endothelial cells culture and analysis exposed to a pulsatile and oscillatory shear stress. Lab Chip. 2009 doi: 10.1039/b909312e. [DOI] [PubMed] [Google Scholar]
- 63.Sei YJ, et al. Detection of frequency-dependent endothelial response to oscillatory shear stress using a microfluidic transcellular monitor. Sci Rep. 2017 doi: 10.1038/s41598-017-10636-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huh D, et al. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci Transl Med. 2012 doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012 doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 66.Stucki AO, Stucki JD, Hall SR, Felder M, Mermoud Y, Schmid RA, Geiser T, Guenat OT. A lung-on-a-chip array with an integrated bio-inspired respiration mechanism. Lab Chip. 2015 doi: 10.1039/c4lc01252f. [DOI] [PubMed] [Google Scholar]
- 67.Marsano A, Conficconi C, Lemme M, Occhetta P, Gaudiello E, Votta E, Cerino G, Redaelli A, Rasponi M. Beating heart on a chip: a novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip. 2016 doi: 10.1039/c5lc01356a. [DOI] [PubMed] [Google Scholar]
- 68.Oh B, et al. Modulating the electrical and mechanical microenvironment to guide neuronal stem cell differentiation. Adv Sci (Weinh) 2021 doi: 10.1002/advs.202002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dai X, et al. Restoration of electrical microenvironment enhances bone regeneration under diabetic conditions by modulating macrophage polarization. Bioact Mater. 2021 doi: 10.1016/j.bioactmat.2020.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo R, et al. Accelerated skin wound healing by electrical stimulation. Adv Healthc Mater. 2021 doi: 10.1002/adhm.202100557. [DOI] [PubMed] [Google Scholar]
- 71.Chen C, et al. Electrical stimulation as a novel tool for regulating cell behavior in tissue engineering. Biomater Res. 2019 doi: 10.1186/s40824-019-0176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu K, et al. Electric fields at breast cancer and cancer cell collective galvanotaxis. Sci Rep. 2020 doi: 10.1038/s41598-020-65566-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weng KC, et al. Human induced pluripotent stem-cardiac-endothelial-tumor-on-a-chip to assess anticancer efficacy and cardiotoxicity. Tissue Eng Part C Methods. 2020 doi: 10.1089/ten.TEC.2019.0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ortega MA, Fernández-Garibay X, Castaño AG, De Chiara F, Hernández-Albors A, Balaguer-Trias J, Ramón-Azcón J. Muscle-on-a-chip with an on-site multiplexed biosensing system for in situ monitoring of secreted IL-6 and TNF-α. Lab Chip. 2019 doi: 10.1039/c9lc00285e. [DOI] [PubMed] [Google Scholar]
- 75.Song S, et al. Collaborative effects of electric field and fluid shear stress on fibroblast migration. Lab Chip. 2013 doi: 10.1039/c3lc41240g. [DOI] [PubMed] [Google Scholar]
- 76.Rifes P, et al. Modeling neural tube development by differentiation of human embryonic stem cells in a microfluidic WNT gradient. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sant S, et al. Biomimetic gradient hydrogels for tissue engineering. Can J Chem Eng. 2010 doi: 10.1002/cjce.20411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ayuso JM, et al. Microfluidic tumor-on-a-chip model to evaluate the role of tumor environmental stress on NK cell exhaustion. Sci Adv. 2021 doi: 10.1126/sciadv.abc2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grant J, et al. Establishment of physiologically relevant oxygen gradients in microfluidic organ chips. Lab Chip. 2022 doi: 10.1039/d2lc00069e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grosberg A, et al. Ensembles of engineered cardiac tissues for physiological and pharmacological study: heart on a chip. Lab Chip. 2011 doi: 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ribas J, Sadeghi H, Manbachi A, Leijten J, Brinegar K, Zhang YS, Ferreira L, Khademhosseini A. Cardiovascular organ-on-a-chip platforms for drug discovery and development. Appl Vitro Toxicol. 2016;1:1. doi: 10.1089/aivt.2016.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kalmykov A, Huang C, Bliley J, Shiwarski D, Tashman J, Abdullah A, Rastogi SK, Shukla S, Mataev E, Feinberg AW, Hsia KJ. Organ-on-e-chip: Three-dimensional self-rolled biosensor array for electrical interrogations of human electrogenic spheroids. Sci Adv. 2019 doi: 10.1126/sciadv.aax0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bavli D, et al. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc Natl Acad Sci USA. 2016 doi: 10.1073/pnas.1522556113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kujala VJ, et al. Laminar ventricular myocardium on a microelectrode array-based chip. J Mater Chem B. 2016 doi: 10.1039/c6tb00324a. [DOI] [PubMed] [Google Scholar]
- 85.Kundu A, et al. Fabrication and characterization of 3D printed, 3D microelectrode arrays for interfacing with a peripheral nerve-on-a-chip. ACS Biomater Sci Eng. 2021 doi: 10.1021/acsbiomaterials.0c01184. [DOI] [PubMed] [Google Scholar]
- 86.Vernekar VN, LaPlaca MC. 3-D multi-electrode arrays detect early spontaneous electrophysiological activity in 3-D neuronal-astrocytic co-cultures. Biomed Eng Lett. 2020 doi: 10.1007/s13534-020-00166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Spira ME, Hai A. Multi-electrode array technologies for neuroscience and cardiology. Nat Nanotechnol. 2013 doi: 10.1038/nnano.2012.265. [DOI] [PubMed] [Google Scholar]
- 88.Adly N, et al. Printed microelectrode arrays on soft materials: from PDMS to hydrogels. NPJ Flexible Electron. 2018;1:1. doi: 10.1038/s41528-018-0027-z. [DOI] [Google Scholar]
- 89.Srinivasan B, et al. TEER measurement techniques for in vitro barrier model systems. J Lab Autom. 2015 doi: 10.1177/2211068214561025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huh D, et al. Reconstituting organ-level lung functions on a chip. Science. 2010 doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Henry OYF, et al. Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab Chip. 2017 doi: 10.1039/c7lc00155j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Renous N, et al. Spatial trans-epithelial electrical resistance (S-TEER) integrated in organs-on-chips. Lab Chip. 2021 doi: 10.1039/d1lc00789k. [DOI] [PubMed] [Google Scholar]
- 93.Weltin A, et al. Cell culture monitoring for drug screening and cancer research: a transparent, microfluidic, multi-sensor microsystem. Lab Chip. 2014 doi: 10.1039/c3lc50759a. [DOI] [PubMed] [Google Scholar]
- 94.Asif A, et al. Microphysiological system with continuous analysis of albumin for hepatotoxicity modeling and drug screening. J Ind Eng Chem. 2021 doi: 10.1016/j.jiec.2021.03.035. [DOI] [Google Scholar]
- 95.Lind JU, et al. Cardiac microphysiological devices with flexible thin-film sensors for higher-throughput drug screening. Lab Chip. 2017 doi: 10.1039/c7lc00740j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Park J, et al. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat Neurosci. 2018 doi: 10.1038/s41593-018-0175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Menon NV, et al. Recapitulating atherogenic flow disturbances and vascular inflammation in a perfusable 3D stenosis model. Biofabrication. 2020 doi: 10.1088/1758-5090/aba501. [DOI] [PubMed] [Google Scholar]
- 98.Lasli S, et al. A human liver-on-a-chip platform for modeling nonalcoholic fatty liver disease. Adv Biosyst. 2019 doi: 10.1002/adbi.201900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Low LA, Mummery C, Berridge BR, Austin CP, Tagle DA. Organs-on-chips: into the next decade. Nat Rev Drug Discov. 2021 doi: 10.1038/s41573-020-0079-3. [DOI] [PubMed] [Google Scholar]
- 100.Picollet-D'hahan N, et al. Multiorgan-on-a-chip: a systemic approach to model and decipher inter-organ communication. Trends Biotechnol. 2021 doi: 10.1016/j.tibtech.2020.11.014. [DOI] [PubMed] [Google Scholar]
- 101.Zhang YS, et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci. 2017 doi: 10.1073/pnas.1612906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Skardal A, et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci Rep. 2017 doi: 10.1038/s41598-017-08879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McAleer CW, Long CJ, Elbrecht D, Sasserath T, Bridges LR, Rumsey JW, Martin C, Schnepper M, Wang Y, Schuler F, Roth AB. Multi-organ system for the evaluation of efficacy and off-target toxicity of anticancer therapeutics. Sci Transl Med. 2019 doi: 10.1126/scitranslmed.aav1386. [DOI] [PubMed] [Google Scholar]
- 104.Xiao S, et al. A microfluidic culture model of the human reproductive tract and 28-day menstrual cycle. Nat Commun. 2017 doi: 10.1038/ncomms14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen HJ, et al. A pumpless body-on-a-chip model using a primary culture of human intestinal cells and a 3D culture of liver cells. Lab Chip. 2018 doi: 10.1039/C8LC00111A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Park D, et al. Integrating organs-on-chips: multiplexing, scaling, vascularization, and innervation. Trends Biotechnol. 2020 doi: 10.1016/j.tibtech.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 107.Wikswo JP, et al. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip. 2013 doi: 10.1039/C3LC50243K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Novak R, et al. Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat Biomed Eng. 2020 doi: 10.1038/s41551-019-0497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Herland A, et al. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat Biomed Eng. 2020 doi: 10.1038/s41551-019-0498-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schuster B, Junkin M, Kashaf SS, Romero-Calvo I, Kirby K, Matthews J, Weber CR, Rzhetsky A, White KP, Tay S. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat Commun. 2020 doi: 10.1038/s41467-020-19058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Puryear Iii JR, et al. Advanced fabrication techniques of microengineered physiological systems. Micromachines. 2020 doi: 10.3390/mi11080730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee S, et al. Modeling 3D human tumor lymphatic vessel network using high-throughput platform. Adv Biol. 2021 doi: 10.1002/adbi.202000195. [DOI] [Google Scholar]
- 113.Sosa-Hernandez JE, et al. Organs-on-a-chip module: a review from the development and applications perspective. Micromachines (Basel) 2018 doi: 10.3390/mi9100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Delon LC, Nilghaz A, Cheah E, Prestidge C, Thierry B. Unlocking the potential of organ-on-chip models through pumpless and tubeless microfluidics. Adv Healthc Mater. 2020 doi: 10.1002/adhm.201901784. [DOI] [PubMed] [Google Scholar]
- 115.Mulholland T, et al. Drug screening of biopsy-derived spheroids using a self-generated microfluidic concentration gradient. Sci Rep. 2018 doi: 10.1038/s41598-018-33055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mohamed MA, Fallahi A, El-Sokkary AM, Salehi S, Akl MA, Jafari A, Tamayol A, Fenniri H, Khademhosseini A, Andreadis ST, Cheng C. Stimuli-responsive hydrogels for manipulation of cell microenvironment: From chemistry to biofabrication technology. Progress Polym Sci. 2019 doi: 10.1016/j.progpolymsci.2019.101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ma Y, Lin M, Huang G, Li Y, Wang S, Bai G, Lu TJ, Xu F. 3D spatiotemporal mechanical microenvironment: a hydrogel-based platform for guiding stem cell fate. Adv Mater. 2018 doi: 10.1002/adma.201705911. [DOI] [PubMed] [Google Scholar]
- 118.Huang G, et al. Functional and biomimetic materials for engineering of the three-dimensional cell microenvironment. Chem Rev. 2017 doi: 10.1021/acs.chemrev.7b00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brown TE, Anseth KS. Spatiotemporal hydrogel biomaterials for regenerative medicine. Chem Soc Rev. 2017 doi: 10.1039/C7CS00445A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lu RXZ, Radisic M. Organ-on-a-chip platforms for evaluation of environmental nanoparticle toxicity. Bioactive Mater. 2021 doi: 10.1016/j.bioactmat.2021.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bhise NS, et al. Organ-on-a-chip platforms for studying drug delivery systems. J Control Release. 2014 doi: 10.1016/j.jconrel.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ahn J, et al. Microfluidics in nanoparticle drug delivery; From synthesis to pre-clinical screening. Adv Drug Deliv Rev. 2018 doi: 10.1016/j.addr.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 123.Guo Y, Luo R, Wang Y, Deng P, Song T, Zhang M, Wang P, Zhang X, Cui K, Tao T, Li Z. SARS-CoV-2 induced intestinal responses with a biomimetic human gut-on-chip. Sci Bull. 2021 doi: 10.1016/j.scib.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Deinhardt-Emmer S, et al. Co-infection with Staphylococcus aureus after primary influenza virus infection leads to damage of the endothelium in a human alveolus-on-a-chip model. Biofabrication. 2020 doi: 10.1088/1758-5090/ab7073. [DOI] [PubMed] [Google Scholar]
- 125.Booth R, Kim H. Characterization of a microfluidic in vitro model of the blood-brain barrier (μBBB) Lab Chip. 2012 doi: 10.1039/C2LC40094D. [DOI] [PubMed] [Google Scholar]
- 126.Kim J, et al. Fungal brain infection modelled in a human-neurovascular-unit-on-a-chip with a functional blood–brain barrier. Nat Biomed Eng. 2021 doi: 10.1038/s41551-021-00743-8. [DOI] [PubMed] [Google Scholar]
- 127.Yadid M, et al. Endothelial extracellular vesicles contain protective proteins and rescue ischemia-reperfusion injury in a human heart-on-chip. Sci Transl Med. 2020 doi: 10.1126/scitranslmed.aax8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Huang D, et al. Reversed-engineered human alveolar lung-on-a-chip model. Proc Natl Acad Sci. 2021 doi: 10.1073/pnas.2016146118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zamprogno P, et al. Second-generation lung-on-a-chip with an array of stretchable alveoli made with a biological membrane. Communications Biology. 2021 doi: 10.1038/s42003-021-01695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ho C-T, et al. Rapid heterogeneous liver-cell on-chip patterning via the enhanced field-induced dielectrophoresis trap. Lab Chip. 2006 doi: 10.1039/B602036D. [DOI] [PubMed] [Google Scholar]