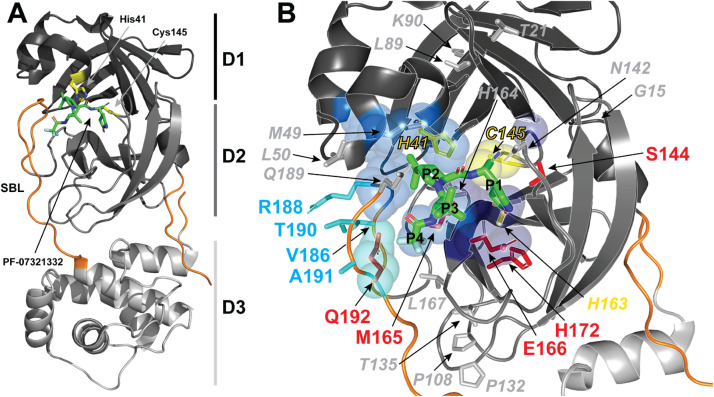
Fig. 1.
The structure of SARS-CoV-2 Mpro in complex with the PF-07321332 inhibitor (PBD 7VH8). A) The Mpro monomer is displayed in cartoon representation to illustrate the domain organization of the protease. Domain I-III (residues 10-99, 100-184, and 201-303, respectively) are colored black, dark grey and light grey, respectively. The substrate binding loop (SBL), amino- and carboxy-termini are colored orange, and PF-07321332 is displayed as green sticks. The catalytic triad (Cys145 and His41) is displayed as yellow sticks. B) Positions of frequent amino acid substitution in Mpro variants are mapped to a detailed view of the contacts between PF-07321332 and Mpro at the interface between Domain I and II. Substrate residue numbers are indicated in black and viral protease amino acids involved in each subsite are displayed as spheres, with S1, S2 and S4 subsites colored dark blue, marine blue and cyan, respectively. Amino acid positions with quantified resistance to nirmatrelvir inhibition, positions that can tolerate multiple mutations without affecting activity or inhibitor binding, positions that yield inactive Mpro, and positions with common variations that are predicted to impart nirmatrelvir resistance are displayed as sticks and colored red, grey, yellow, and cyan, respectively.