Figure 1. METTL3 interacts with p53 and enhances p53 protein accumulation.
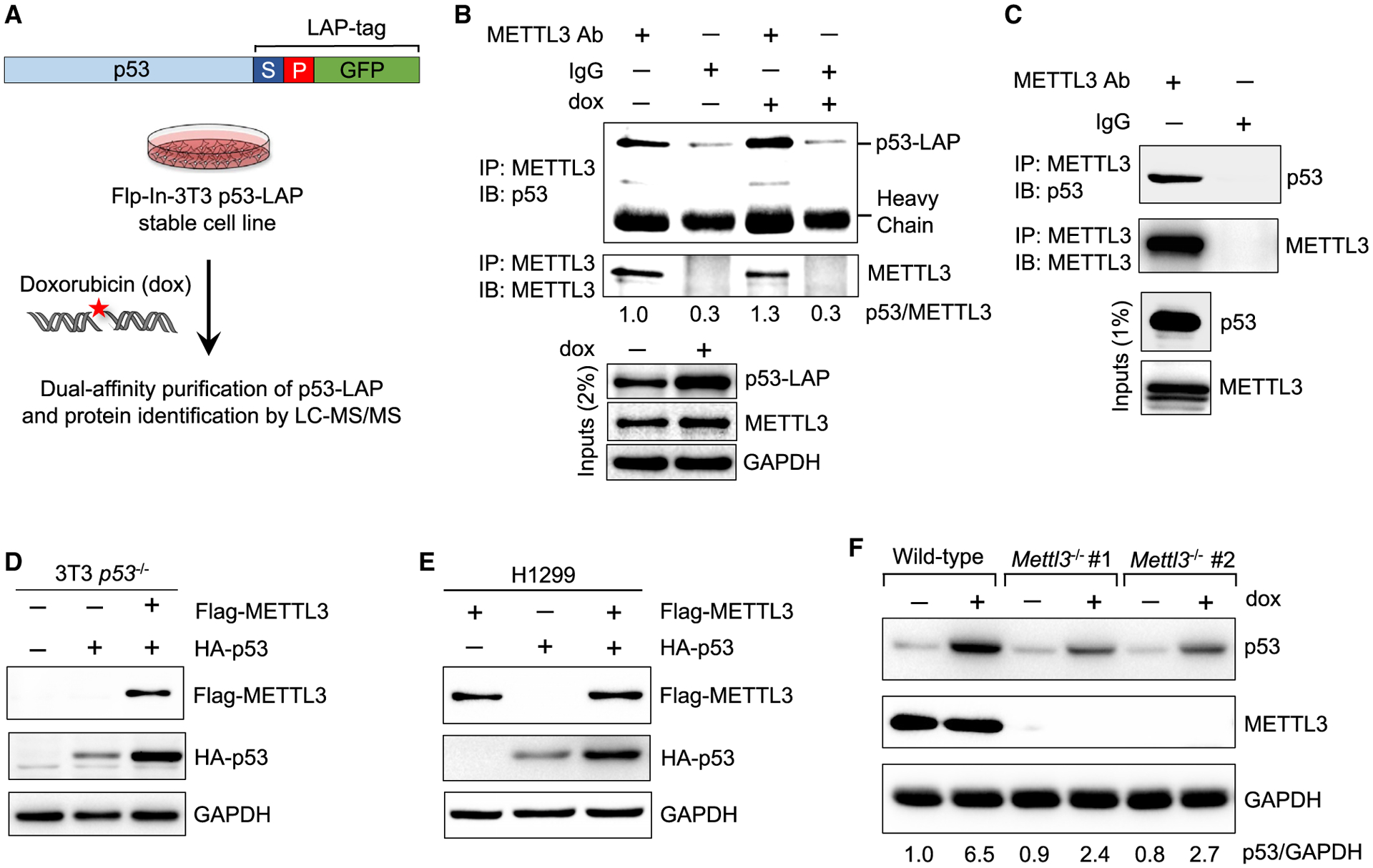
(A) Schematic of dual-affinity purification of LAP-tagged p53 protein in Flp-In-3T3 fibroblasts. Cells were treated with 0.2 μg/mL doxorubicin (dox) for 6 h, followed by dual-affinity purification of p53-bound protein complexes and protein identification by LC-MS/MS.
(B) Co-immunoprecipitation (coIP) and immunoblot assay to examine p53-LAP and endogenous METTL3 interaction in untreated and DNA damage-treated Flp-In-3T3 cells. Numbers indicate the amount of p53 co-precipitated relative to METTL3 (n = 2).
(C) CoIP and immunoblot assay to examine interaction of endogenous METTL3 and p53 in E1A;HRasG12V-expressing MEFs (n = 2).
(D) Immunoblot after transfection of FLAG-METTL3 and HA-p53 plasmids into Flp-In-3T3 p53−/− cells (n = 2).
(E) Immunoblot after co-transfection of FLAG-METTL3 and HA-p53 plasmids into H1299 cells (n = 3).
(F) p53 induction after 6 h dox (0.2 μg/mL) in wild-type (WT) and two different Mettl3−/− ES cell lines. Numbers indicate the amount of p53 relative to glycer-aldehyde 3-phosphate dehydrogenase (abbreviated GAPDH) loading control (n = 3). Representative immunoblots are shown in (B)–(F), and GAPDH serves as a loading control.
See also Figures S1 and S2A and Table S1.
