Abstract
The global COVID-19 pandemic, caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was first detected in China in December 2019. To date, there have been approximately 3.4 million reported cases of COVID-19 and over 24,000 deaths in Thailand. In this study, we investigated the molecular characteristics and evolution of SARS-CoV-2 in Thailand from 2020 to 2022. Two hundred sixty-eight SARS-CoV-2 isolates, collected mostly in Bangkok from COVID-19 patients, were characterised by partial genome sequencing. Moreover, the viruses in 5,627 positive SARS-CoV-2 samples were identified as viral variants – B.1.1.7 (Alpha), B.1.617.2 (Delta), B.1.1.529 (Omicron/BA.1), or B.1.1.529 (Omicron/BA.2) – by multiplex real-time reverse transcription polymerase chain reaction (RT-PCR) assays. The results revealed that B.1.36.16 caused the predominant outbreak in the second wave (December 2020–January 2021), B.1.1.7 (Alpha) in the third wave (April–June 2021), B.1.617.2 (Delta) in the fourth wave (July–December 2021), and B.1.1.529 (Omicron) in the fifth wave (January–March 2022). The evolutionary rate of the viral genome was 2.60 × 10-3 (95% highest posterior density [HPD], 1.72 × 10-3 to 3.62 × 10-3) nucleotide substitutions per site per year. Continued molecular surveillance of SARS-CoV-2 is crucial for monitoring emerging variants with the potential to cause new COVID-19 outbreaks.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00705-022-05666-6.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection caused the COVID-19 pandemic as declared by the World Health Organization (WHO) on 11 March 2020 and continues to impact public health and world economies [1]. As of 14 September 2022, over 600 million confirmed COVID-19 cases were reported, with over 6 million fatalities globally [2]. SARS-CoV-2 is an enveloped virus with a positive-sense, single-stranded RNA genome of approximately 30 kb [3]. The genome encodes four structural proteins (spike [S], envelope [E], membrane [M], and nucleocapsid [N]) and 16 non-structural proteins (nsp1-nsp16), which are involved in viral function and replication. The massive circulation of SARS-CoV-2 worldwide and inequitable global vaccine distribution has led to evolutionary pressure on the virus and the emergence of new variants [4, 5]. An abundance of SARS-CoV-2 genome sequences has been generated rapidly and deposited in the global archive, namely the Global Initiative on Sharing All Influenza Database (GISAID). As of 2 March 2021, significant mutations in the viral genome led to the classification of variants into nine clades (S, L, V, G, GH, GK, GR, GV, and GRY) [6]. The epidemiologically relevant phylogenetic cluster of SARS-CoV-2 is further defined as a lineage by the Phylogenetic Assignment of Named Global Outbreak Lineages (PANGOLIN) tool [7]. Based on their enhanced transmissibility, increased virulence, and decreased susceptibility to natural-infection- and vaccine-mediated neutralizing antibodies attributed to significant amino acid substitutions, several SARS-CoV-2 variants have been classified as Variants of Concern (VOCs) [8]. To assist in public communication and to avoid stigmatisation, letters of the Greek alphabet, i.e., Alpha, Beta, Gamma, Delta, and Omicron, are used to designate SARS-CoV-2 variants [8].
The first confirmed COVID-19 case in Thailand was reported on 12 January 2020 in a traveller from China [9]. The number of infected individuals surged rapidly during March–May 2020 because of transmission linked to boxing events and entertainment venues in Bangkok, and the virus then spread throughout Thailand, and this was considered the first COVID-19 wave [10]. To effectively contain spread of the virus, public health and social measures, including wearing masks, physical distancing, movement restriction, workplace and school closures, and city lockdowns, were implemented. On 17 December 2020, Thailand entered the second wave of the COVID-19 epidemic, with reported daily cases of over 1,500, triggered by spread among migrants working at the Central Shrimp Market, Samut Sakhon Province [11]. The third wave began in early April 2021 with the upsurge of COVID-19 cases linked to an entertainment venue in the Thonglor district of Bangkok [12]. This severe and deadly wave was driven by the emergence of a more transmissible B.1.1.7 (Alpha) SARS-CoV-2 VOC, leading to rising hospitalisations and overwhelming healthcare facilities [13]. With the exception of the city lockdown, all protective measures implemented by the government were still in effect at these times. Moreover, field hospitals were set up to handle patient isolation. Amid the third wave, a mass vaccination campaign was rolled out on 7 June 2021 to slow down transmission. However, the supply of vaccines was limited, and only two vaccines, CorovaVac and Vaxzevria, were available at that time. As of 1 July 2021, there were 52,052 confirmed COVID-19 cases with 1,971 patients classified as having severe illness, 566 of which required ventilator support [14]. As of 8 July 2021, COVID-19 cases were reported in all 77 provinces of Thailand. The Centre for COVID-19 Situation Administration (CCSA) declared the emergence of a fourth wave of the COVID-19 pandemic in Thailand caused by the highly contagious Delta variant, whose transmissibility was faster than that of the previous SARS-CoV-2 variant [14]. By December 2021, Thailand experienced its fifth COVID-19 wave, with approximately 3.4 million confirmed COVID-19 cases and over 24,000 deaths as of the end of March 2022 [15].
In our previous molecular epidemiological investigation of SARS-CoV-2 in Thailand during the first wave of the outbreak in 2020, 40 nasopharyngeal and/or throat swab specimens were found to contain SARS-CoV-2 types L, GH, GR, O, and S [16]. In this study, we monitored and tracked emerging new variants of SARS-CoV2 circulating in Thailand between March 2020 and March 2022. We found epidemiological patterns of SARS-CoV-2 infection in Thailand that could have implications for more-effective disease surveillance and public health preparedness.
Materials and methods
Sample collection and RNA extraction
All nasopharyngeal swab samples were collected as part of outbreak investigations during the period of the first through fifth waves and state quarantine (SQ) from March 2020 to March 2022 (Fig. 1). The SQ samples were from Thai citizens and foreigners who were required to stay in government-approved facilities for 14 days. Prior to analysis in our laboratory, total nucleic acid was extracted from 200 μL of supernatant using a magLEAD 12gC instrument (Precision System Science, Chiba, Japan) according to the manufacturer’s instructions.
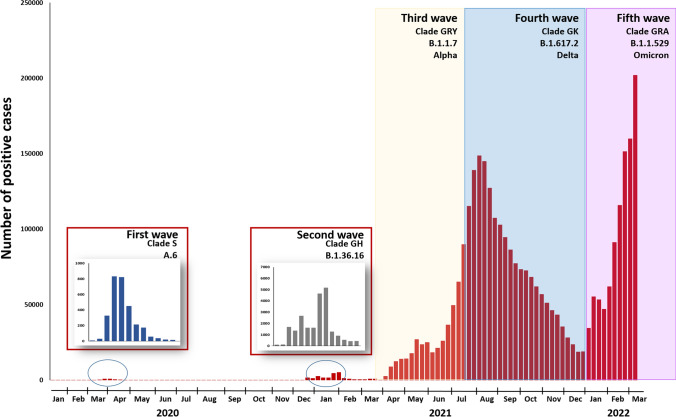
Fig. 1.
Timeline showing the SARS-CoV-2 epidemic wave in Thailand, 2020-2022 [9]
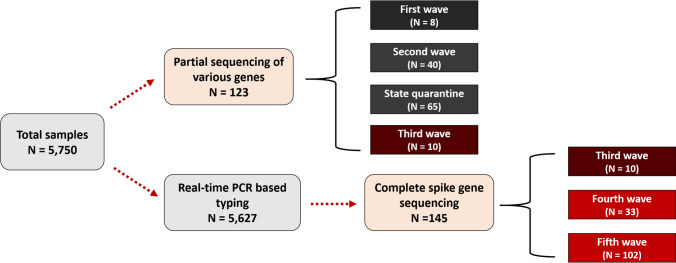
During the study, 5,750 selected nasopharyngeal swab samples submitted to the collaborating hospitals and the Institute of Urban Disease Control and Prevention (IUDC) tested positive for SARS-CoV-2 by multiplex real-time reverse transcription polymerase chain reaction (RT-PCR) assays described earlier [16]. Due to the small outbreaks and high similarity of SARS-CoV-2 variants during first and second epidemic waves, including SQ, 123 samples were randomly selected for partial sequencing of various genomes (Fig. 2). Between the third and fifth epidemic waves, 5,627 samples were subsequently typed by multiplex real-time RT-PCR for rapid simultaneous typing of SARS-CoV-2 variants from the large-scale outbreak. Among them, 145 entire spike genes were successfully sequenced from individual COVID-19 patients.
Fig. 2.
Flow chart of genotyping of SARS-CoV-2 strains during the study period
Genome sequencing
Complete spike gene sequences (nucleotide position 21,346–25,468) and partial sequences of ORF1ab (nucleotide position 8,596–8,943 and 15,074–16,269), ORF3a to E (nucleotide position 25,017–25,639 and 25,903-26,278), and ORF8 to N (nucleotide position 28,147–29,041) were determined using a SuperScript III Platinum One-Step RT-PCR System (Invitrogen, Carlsbad, CA, USA) and 11 sets of oligonucleotide primers (Supplementary Table S1). Briefly, the RT-PCR reactions were performed in a total volume of 25 μL containing 2–3 μL of 100 ng to 1 µg total RNA, 0.5 μM each primer, 12.5 μL of 2X reaction mix (containing 0.4 mM each dNTP and 3.2 mM MgSO4), 1 μL of SSIII RT/Platinum Taq Mix, and nuclease-free water. The conventional RT-PCR was performed using a thermal cycler (Eppendorf, Hamburg, Germany). Cycling conditions included a reverse transcription step at 45 °C for 30 min, an initial denaturation step at 94 °C for 3 min in order to activate the Platinum Taq DNA Polymerase, 40 cycles of amplification consisting of 30 s of denaturation at 94 °C, 30 s of primer annealing at 53 °C, and 90 s of extension at 68 °C, followed by further extension for 7 min at 68 °C. The PCR amplicons were separated on a 2% agarose gel with a 100-base pair DNA ladder and visualized on an ultraviolet transilluminator. The amplified products from the PCR reactions were purified using a HiYield Gel/PCR DNA Fragment Extraction Kit (RBC Bioscience Co, Taipei, Taiwan) according to the manufacturer's specifications. The purified products were sequenced by First BASE Laboratories Sdn Bhd, Selangor, Malaysia, and the nucleotide sequences were deposited in the GenBank database under the accession numbers OK083891-OK084640, OM984745-OM984850, and OM996047-OM996083 (Supplementary Table S2).
Multiplex real-time RT-PCR assay
Primers and probes specific for B.1.1.7 (Alpha), B.1.617.2 (Delta), B.1.1.529 (Omicron/BA.1), and B.1.1.529 (Omicron/BA.2) were designed to target the S gene. The sequences of primers and probes were selected from conserved regions of sequences available in the GISAID database (http://www.gisaid.org/). The multiplex primer sets and TaqMan probes are shown in Supplementary Table S3. Primers and probes were used at a final concentration of 0.5 and 0.25 µM, respectively. A combination of 3.0 µl of 100 ng to 1 µg total RNA with a reaction mixture containing 10 µl of 2× SensiFAST Probe One-Step mix, 0.2 µl of reverse transcriptase, and 0.4 µl of RiboSafe RNase Inhibitor, 1.25 mM MgCl2, 0.25 mM dNTPs, and RNase-free water was used in a final volume of 20 µl. One-step multiplex real-time RT-PCR was performed using LightCycler 480 real-time PCR system (Roche, Mannheim, Germany). The thermocycling conditions included a reverse transcription step at 42°C for 30 min and a hot start DNA Taq polymerase activation step at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 30 s. Multiple fluorescent signals were obtained once per cycle upon completion of the extension step. Data acquisition and analysis of the real-time PCR results were performed using LightCycler 480 SW1.5 software (Roche).
Samples that had previously been identified as B.1.1.7 (Alpha), B.1.617.2 (Delta), B.1.1.529 (Omicron/BA.1), and B.1.1.529 (Omicron/BA.2) served as the controls for the one-step multiplex real-time RT-PCR assay. Plasmids were constructed by insertion of the spike genes of B.1.1.7 (Alpha) (nt 21711–21860; SARS-CoV-2/human/THA/CU490/2020, OK084567), B.1.617.2 (Delta) (nt 21962–22082; SARS-CoV-2/human/THA/CU2750/2021, OK084639), B.1.1.529 (Omicron/BA.1) (nt 22174–22274; SARS-CoV-2/human/THA/Spike_CU6883/2022, OM984777), and B.1.1.529 (Omicron/BA.2) (nt 21595–21729; SARS-CoV-2/human/THA/Spike_CU7559/2022, OM984826) into pGEM-T Easy Vector (Promega, Madison, WI), using a TA-cloning strategy.
Phylogenetic analysis
The sequence datasets were constructed using BioEdit v7.2.6 software [17] and aligned using CLUSTAL W on the European Bioinformatics Institute (EBI) webserver [18].
The diversity of SARS-CoV-2 lineages was analysed by the maximum-likelihood (complete spike gene) and the neighbor-joining (partial genes) phylogenetic methods available in the MEGA program (v7) [19]. For phylogenetic trees, the best-fit nucleotide substitution model (Tamura 3-parameter with gamma distribution) was selected according to the Bayesian information criterion (BIC) using the likelihood ratio test as implemented in MEGA. The bootstrap method was used to determine the statistical consistency of the tree nodes (1000 random samplings).
A time-scaled phylogenetic tree for the complete spike gene was constructed using BEAST version 1.10.4 [20]. For Bayesian phylogenetic analysis, an uncorrelated log-normal prior distribution of nucleotide substitution rates among lineages was used. The general time-reversible (GTR) model was selected as the nucleotide substitution model. The nucleotide substitution rate and time to most recent common ancestor (TMRCA) were calculated for the spike gene using the Bayesian Markov chain Monte Carlo (BMCMC) method as implemented in the program BEAST [20]. Bayesian Markov chain Monte Carlo analysis was run for 100 million steps, 10% of which were removed as burn-in and sampled every 1,000 steps from the posterior distribution. Tracer version 1.7.1 tool (http://tree.bio.ed.ac.uk/software/tracer/) was used to assess for the convergence of all parameters (operator effective sample size of > 200). A maximum-clade-credibility (MCC) tree was constructed using the TreeAnnotator v1.10.4 tool (http://beast.bio.ed.ac.uk/treeannotator).
Results
Distribution of SARS-CoV-2 outbreaks in Thailand
From March 2020 to June 2021, 123 confirmed cases (first wave, N = 8; second wave, N = 40; third wave, N = 10; state quarantine, N = 65) were successfully genotyped by partial SARS-CoV-2 genome sequencing. In this study, the first wave of the outbreak (March–May 2020) in Thailand was characterised by two different lineages, A and B.1 (Fig. 1 and Supplementary Fig. S1). During the period of SQ from May 2020 through May 2021, we received 65 clinical specimens obtained from travellers and Thais who presented with or without symptoms of COVID-19 and were admitted to a hospital or hotel in Bangkok or Chon Buri province. Among these, lineage B.1.1 was the most frequently detected genotype and accounted for 29.2% of the isolates (19/65), followed by 48% (31/65) for lineage B.1. Of the remaining isolates, 11 were classified as lineage B.1.1.7 (Alpha), and another three were of lineage B.1.177 (Supplementary Fig. S1 and Supplementary Table S2). The strains from SQ were imported from the Americas (12.3%), Asia (41.5%), Europe (23.1%), and unknown (23.1%). The results showed that lineage B.1.1.7 (Alpha) was imported from the United States, France, Slovenia, and the United Kingdom (UK). Lineage B.1.177 was imported from the UK and the United Arab Emirates (UAE). Lineage B.1.1 was predominantly imported from Asian countries (Qatar, India, the Philippines, Japan, and Bahrain), the UK, and Italy. Lineage B.1 was imported from Asian and European countries. During the second (October 2020–March 2021) and the third (April 2021–June 2021) waves of the outbreak, lineage B.1 and B.1.1.7, respectively, became the predominant virus.
Multiplex real-time RT-PCR assays to differentiate variants of SARS-CoV-2
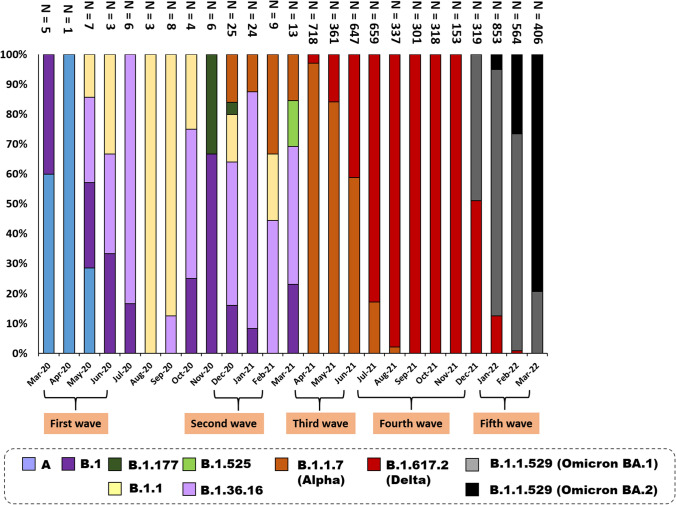
In Thailand, predominant variants were detected in different epidemic waves (Fig. 3). From March 2020 to 14 March 2022, the viruses in 5,627 samples were identified as B.1.1.7 (Alpha), B.1.617.2 (Delta), or B.1.1.529 (Omicron BA.1 and BA.2) using multiplex real-time RT-PCR. The results showed that clade B.1.1.7 (Alpha) was the most frequent variant in the third epidemic wave (1,510/5,627: 26.8%). B.1.617.2 (Delta) (2,382/5,627: 42.3%) was the predominant strain responsible for SARS-CoV-2 infection in the fourth epidemic wave. B.1.1.529 (Omicron BA.1) was first detected in Thailand in mid-December 2021 (1,375/5,627: 24.4%), which caused a nationwide epidemic wave until the end of February 2022. Since then, clade B.1.1.529 (Omicron BA.2) emerged in Thailand at the end of January 2022 (360/5,627: 6.4%) and became the major variant in early March 2022.
Fig. 3.
The time course of variant distribution in Thailand during 2020-2022
In this study, the genotyping results obtained by multiplex real-time RT-PCR were identical to those obtained by nucleotide sequencing (N = 268), indicating that the SARS-CoV-2 variants had been accurately genotyped using the multiplex real-time RT-PCR assay.
Phylogenetic relationships
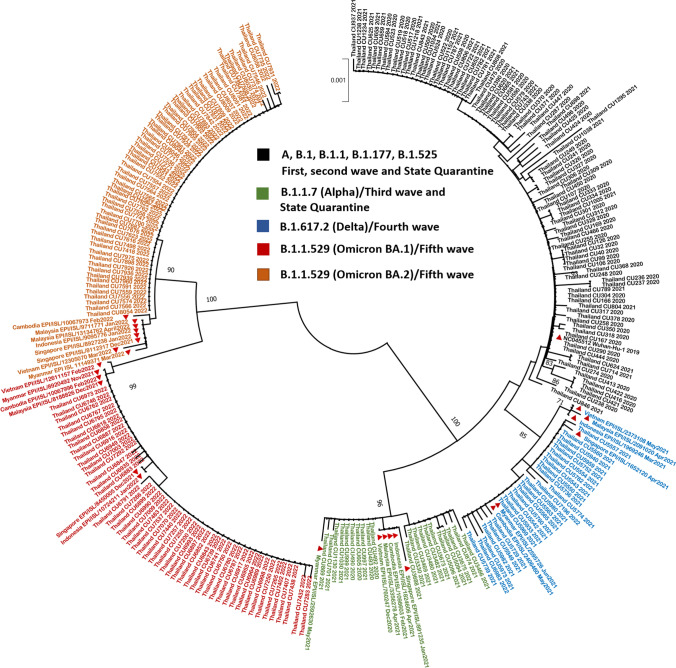
The nucleotide sequences of the complete spike genes of 155 SARS-CoV-2 isolates were determined to evaluate the genetic relationships among Thai SARS-CoV-2 strains. These 155 isolates, belonging to B.1.1.7 (Alpha) (N = 20), B.1.617.2 (Delta) (N = 33), B.1.1.529 (Omicron BA.1) (N = 48), and B.1.1.529 (Omicron BA.2) (N = 54), were identified by amplification of the partial S gene, which harbours the major antigenic sites in SARS-CoVs. Sequences from 123 samples collected between the first and third epidemic waves and SQ were also included. The phylogenetic tree based on S sequences showed that the members of clade B.1.1.529 (Omicron BA.1 and BA.2) clustered together and were differentiated from the other clades with bootstrap values >95% (Fig. 4).
Fig. 4.
Maximum-likelihood phylogenetic analysis based on the complete nucleotide sequences of the spike genes of SARS-CoV-2 isolates from Thai patients during 2020-2022 and those from other Southeast Asian countries (red triangles). The scale bar indicates the number of nucleotide substitutions per site.
The S gene analysis showed that the B.1.1.7 (Alpha) differed from the Wuhan-Hu1 strain by seven amino acid substitutions: N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H. The B.1.177 variants contained the mutations L16F, D215H, A222V, N370S, and D614G. Several mutations were identified in the sequences of B.1.617.2 (Delta) variants, namely, T17R, T93I, G140D, L452R, T478K, D614G, P681R, and D950N. Forty-two and 31 amino acid substitutions were detected in the B.1.1.529 (Omicron BA.1) and the B.1.1.529 (Omicron BA.2) isolates, respectively. In addition, one strain (Thailand_CU8056) that was clustered in the B.1.1.529 (Omicron BA.2) clade carried an I1221T substitution in the S protein (Fig. 4).
The 123 partial sequences of SARS-CoV-2 identified in this study were also analysed for mutations compared with the Wuhan-Hu1 strain. Nineteen amino acid substitutions were identified, as shown in Supplementary Table S4, and these mutations were distributed across four genes of the SARS-CoV-2 genome. These included two changes in ORF1b gene, three changes in the ORF3a gene, 10 changes in the N gene, and a single change in ORF8 gene.
The evolutionary history of the structural region of SARS-CoV-2 was investigated by performing Bayesian analysis with a SARS-CoV-2 S glycoprotein sequence data set. The mean evolution rate was 2.60 × 10-3 (95% highest posterior density [HPD], 1.72 × 10-4 to 3.62 × 10-4) substitutions per site per year (Supplementary Fig. S2). The most recent common ancestor of all SARS-CoV-2 clades dated to September 2019.
Discussion
SARS-CoV-2 has been circulating in Thailand since early 2020. The SARS-CoV-2 fourth wave outbreak is the largest known in Thailand, with over 800,000 recorded cases at the end of December 2021 [15]. In this study, we determined the partial genome sequences of 268 Thai strains, performed phylogenetic analysis, and analysed their molecular evolution to investigate their relationships to previously described viruses. The first outbreak began in Thailand in 2020, with the first imported case occurring in late January 2020, and spread to several provinces, with Bangkok being the most severely affected [9]. This outbreak was attributed to lineage A.6 (S) variants, which were responsible for 67.5% of all SARS-CoV-2 cases [16]. This study showed that clade GH rapidly became the predominant variant throughout Thailand during the second wave of the COVID-19 epidemic (late December 2020). In November 2020, the B.1.1.7 (Alpha) variant emerged for the first time in the United Kingdom and caused higher mortality [21]. The B.1.1.7 (Alpha) variant subsequently spread through Europe, the United States, and Asia over the next two months. Between April and June 2021, a third wave of the B.1.1.7 (Alpha) variant peaked in Thailand. The emergence of the B.1.617.2 (Delta) variant in India in October 2020 caused large-scale outbreaks on that subcontinent [22], and it became the dominant variant worldwide by February 2021. In Thailand, the fourth epidemic peak was observed during August-September 2021, mainly caused by the B.1.617.2 (Delta) variant. The B.1.1.529 (Omicron) variant was first identified in South Africa in November 2021 [23]. After its emergence, the B.1.1.529 (Omicron) variant replaced the B.1.617.2 (Delta) variant, and it has circulated as the dominant variant in several countries since December 2021. In mid-December 2021, the B.1.1.529 (Omicron) variant was detected in Thailand. The largest number of COVID-19 cases occurred in March 2022, when COVID-19 case numbers surged, mainly due to this variant.
A study in Malaysia showed that lineage B.6 (O)-associated groups and B.1.524 (G) were the predominantly detected variants throughout the country during the second (27 February–8 July 2020) and third (8 October 2020) epidemic wave, respectively [24]. During the same period in Thailand, most strains were classified as the lineage A.6 or B.1.36.16. In Vietnam, there was a reported increase in two clusters of SARS-CoV-2 as the waves of virus infection progressed between July 2020 and February 2021, and the major causative agent was lineage B.1.1 with a novel mutation in nsp9 [25]. According to the results of this study, the most prevalent variant in Thailand during July 2020 to February 2021 was lineage B.1.36.16.
Several strains with mutations in the S protein are variants of concern (VOCs) with potentially enhanced transmissibility and infectivity [26, 27]. The D614G mutation in the S protein, which increases the ability of the virus to replicate in the upper respiratory tract, causes a possible conformational change in the S1 subunit and increases furin cleavage efficiency at the S1/S2 site [28, 29]. The D614G mutation in the S protein was first detected outside of China in a small outbreak in Germany in January 2020 [30]. Our analysis showed that almost 90% of the Thai variants contained the D614G mutation. The spike mutations N501Y and K417N were first recorded in the B.1.1.7 (Alpha) and B.1.351 (Beta) variant, respectively; the presence of these substitutions in the receptor-binding domain (RBD) of the S protein confers increased binding affinity of the virus to the ACE2 receptor [31–33]. The results of this study also showed that the N501Y mutation was present in 100% of the B.1.1.7 (Alpha) isolates. As reported recently, the E484K substitution was first detected in the B.1.351 (Beta) variants and has been associated with antibody neutralisation escape by directly reducing antibody binding affinity [34, 35]. An analysis of the B.1.617.2 (Delta) variant showed that the variant contained a T-to-G transversion at nucleotide position 22,917, resulting in an L452R mutation in the S protein, which is also found in the B.1.617.1 (Kappa) and B.1.427 and B.1.429 (Epsilon) variants. However, no B.1.351 (Beta), B.1.617.1 (Kappa), or B.1.429 (Epsilon) isolates were found in the present study. The L452R substitution is located in the RBD of the spike protein and results in a reduction in antibody neutralising activity [36]. The spike-L452R substitution was consistently observed in this study in the B.1.617.2 (Delta) variant.
In this study, we detected several mutations that occurred in different regions of the partial sequences. The mutation Q57H in the ORF3a gene has been found in Indian and global variants when compared to the Wuhan-Hu-1, and it plays a potential role in viral pathogenesis [37]. This amino acid substitution was observed in all samples belonging to the lineage B.1.36.16 in this study. The mutation S194L in the N gene was observed in the majority of deceased patients from India [37]. All of the lineage B.1.36.16 isolates from the second epidemic wave in the current study contained this substitution.
Our study showed that the mean evolutionary rate early in the epidemics was 2.60 × 10-3 nucleotide substitutions per site per year. This rate is approximately four times as high as that reported in Pakistan, 5.68 × 10-4 substitutions per site per year [38]. Our estimate was similar to those of previously published reports (0.99 – 1.8 × 10-3 substitutions per site per year) [39–42].
A limitation of this study is that partial genome sequencing data were not available for all outbreak samples, since S region typing is not yet routinely performed in our laboratory. We successfully obtained SARS-CoV-2 typing data by using a multiplex real-time RT-PCR assay between the third and fifth epidemic waves, occurring primarily in the last year of the study. The present study highlights the importance of molecular typing for a complete understanding of the diversity and circulation of SARS-CoV-2.
In summary, a SARS-CoV-2 outbreak has been ongoing in Thailand for more than two years, with a total of five epidemic waves. Clade B.1.36.16 (GH) predominated in the second epidemic wave (2021), clade B.1.1.7 (Alpha) in the third wave (2021), clade B.1.617.2 (Delta) in the fourth wave (2021), and clade B.1.1.529 (Omicron) in the fifth wave (2022), indicating that new epidemic waves occurred due to emerging strains. Continued molecular surveillance of SARS-CoV-2 is crucial for monitoring emerging variants to prevent possible new COVID-19 outbreaks.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This study was funded by the National Research Council of Thailand (NRCT), the Health Systems Research Institute (HSRI), the Center of Excellence in Clinical Virology at Chulalongkorn University, King Chulalongkorn Memorial Hospital, and the Rachadapisek Sompote Fund of Chulalongkorn University for postdoctoral fellowships to Jiratchaya Puenpa.
Author contributions
J.P. and P.R. analysed data and drafted the manuscript. P.R., N.S., and J.C. performed the laboratory experiments and collected data. R.Y. and N.S. collected specimens. K.S., S.T., S.V., and Y.P. supervised the study and finalized the manuscript. All authors reviewed the manuscript.
Data availability
The datasets generated during the current study are available in the GenBank database (https://www.ncbi.nlm.gov/genome) under accession numbers OK083891-OK084640, OM984745-OM984850, and OM996047-OM996083.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics statement
The study protocol was approved by the Institutional Review Board of the Faculty of Medicine of Chulalongkorn University (IRB number 178/64). A waiver of the requirement for written informed consent was granted by the institutional review board of the Ethics Committee for Human Research due to the anonymity of the samples. All procedures were performed in accordance with the relevant guidelines and regulations.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO (2020) Coronavirus disease 2019 (COVID-19). Situation Report-51. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200311-sitrep-51-covid-19.pdf. Accessed 22 Mar 2022
- 2.WHO (2021) Weekly epidemiological update on COVID-19 – 14 September 2022. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---14-september-2022. Accessed 19 Sept 2022
- 3.Wang MY, Zhao R, Gao LJ, Gao XF, Wang DP, Cao JM. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. 2020;10:587269. doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad L. Implication of SARS-CoV-2 immune escape spike variants on secondary and vaccine breakthrough infections. Front Immunol. 2021;12:742167. doi: 10.3389/fimmu.2021.742167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bui NN, Lin YT, Huang SH, Lin CW. Haplotype distribution of SARS-CoV-2 variants in low and high vaccination rate countries during ongoing global COVID-19 pandemic in early 2021. Infect Genet Evol. 2022;97:105164. doi: 10.1016/j.meegid.2021.105164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.GISAID (2021) Clade and lineage nomenclature aids in genomic epidemiology studies of active hCoV-19 viruses. https://www.gisaid.org/references/statements-clarifications/clade-and-lineage-nomenclature-aids-in-genomic-epidemiology-of-active-hcov-19-viruses/. Accessed 5 Feb 2022
- 7.Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, du Plessis L, Pybus OG. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO (2021) Tracking SARS-CoV-2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/. Accessed 15 Jan 2022
- 9.WHO (2020) Coronavirus disease 2019 (COVID-19). Situation Report-1. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf. Accessed 5 Jan 2022
- 10.WHO (2020) Coronavirus disease 2019 (COVID-19). Thailand Situation Report. https://www.who.int/docs/default-source/searo/thailand/2020-03-15-tha-sitrep-22-covid19.pdf. Accessed 20 Feb 2022
- 11.WHO (2021) Coronavirus disease 2019 (COVID-19). Thailand Situation Report-109. https://cdn.who.int/media/docs/default-source/searo/thailand/2020_12_30_tha-sitrep-110-covid19.pdf?sfvrsn=fb123e61_5. Accessed 1 Oct 2022
- 12.WHO (2021) Coronavirus disease 2019 (COVID-19). Thailand Situation Report-167. https://cdn.who.int/media/docs/default-source/searo/thailand/2021_04_05_eng_sitrep-167-covid19.pdf?sfvrsn=8b63d890_5. Accessed 20 Dec 2021
- 13.WHO (2021) Coronavirus disease 2019 (COVID-19). Thailand Situation Report-170. https://cdn.who.int/media/docs/default-source/searo/thailand/2021_04_09_eng_sitrep_170-covid19.pdf?sfvrsn=eee4c530_8. Accessed 20 Dec 2021
- 14.WHO (2021) Coronavirus disease 2019 (COVID-19). Thailand Situation Report-191. https://cdn.who.int/media/docs/default-source/searo/thailand/2021_07_08_eng-sitrep-191-covid19-revision.pdf?sfvrsn=7c57b84_3. Accessed 20 Dec 2021
- 15.Ministry of public health (MOPH), Thailand (2022) Coronavirus disease 2019 (COVID-19) WHO Thailand Situation. https://ddc.moph.go.th/covid19-dashboard/ (In Thai). Accessed 15 Jan 2022
- 16.Puenpa J, Suwannakarn K, Chansaenroj J, Nilyanimit P, Yorsaeng R, Auphimai C, Kitphati R, Mungaomklang A, Kongklieng A, Chirathaworn C, Wanlapakorn N, Poovorawan Y. Molecular epidemiology of the first wave of severe acute respiratory syndrome coronavirus 2 infection in Thailand in 2020. Sci Rep. 2020;610:16602. doi: 10.1038/s41598-020-73554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 18.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Public Health England (2021) Coronavirus disease 2019 (COVID-19). Variants: distribution of cases data. https://www.gov.uk/government/publications/covid-19-variants-genomically-confirmed-case-numbers/variants-distribution-of-cases-data. Accessed 1 June 2022
- 22.Callaway E. Delta coronavirus variant: scientists brace for impact. Nature. 2021;595:17–18. doi: 10.1038/d41586-021-01696-3. [DOI] [PubMed] [Google Scholar]
- 23.Poudel S, Ishak A, Perez-Fernandez J, Garcia E, León-Figueroa DA, Romaní L, Bonilla-Aldana DK, Rodriguez-Morales AJ. Highly mutated SARS-CoV-2 Omicron variant sparks significant concern among global experts—what is known so far? Travel Med Infect Dis. 2022;45:102234. doi: 10.1016/j.tmaid.2021.102234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan KK, Tan JY, Wong JE, Teoh BT, Tiong V, Abd-Jamil J, Nor'e SS, Khor CS, Johari J, Yaacob CN, Zulkifli MM, CheMatSeri A, Mahfodz NH, Azizan NS, AbuBakar S. Emergence of B.1.524(G) SARS-CoV-2 in Malaysia during the third COVID-19 epidemic wave. Sci Rep. 2021;11:22105. doi: 10.1038/s41598-021-01223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phuong HVM, Tung TS, Trang UTH, Hang NLK, Son NV, Hien PT, Thanh LT, Cuong VD, Thanh TT, Nhan NTT, Duong TN, Nghia ND, Tu TA, Choisy M, Rabaa MA, Rogier van Doorn H, Anh DD, Mai LQ. Novel mutation of SARS-CoV-2, Vietnam, July 2020. Emerg Infect Dis. 2021;27:1519–1521. doi: 10.3201/eid2705.210013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yurkovetskiy L, Wang X, Pascal KE, et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang H, Gao L, Wu Z, Meng F, Zhao X, Shao Y, Shi X, Qiao S, An J, Du X, Qin FX. Characterization of SARS-CoV-2 variants N501Y.V1 and N501Y.V2 spike on viral infectivity. Front Cell Infect Microbiol. 2021;11:720357. doi: 10.3389/fcimb.2021.720357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plante JA, Liu Y, Liu J, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Manne K, Stalls V, Kopp MF, Henderson R, Edwards RJ, Haynes BF, Acharya D614G mutation alters SARS-CoV-2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep. 2021;34:108630. doi: 10.1016/j.celrep.2020.108630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Böhmer MM, Buchholz U, Corman VM, et al. Investigation of a COVID-19 outbreak in Germany resulting from a single travel-associated primary case: a case series. Lancet Infect Dis. 2020;20:920–928. doi: 10.1016/S1473-3099(20)30314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, Dingens AS, Navarro MJ, Bowen JE, Tortorici MA, Walls AC, King NP, Veesler D, Bloom JD. Deep mutational scanning of SARS-CoV-2 receptor bindingdomain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makowski L, Olson-Sidford W, W-Weisel J, Biological and clinical consequences of integrin binding via a rogue RGD motif in the SARS CoV-2 spike protein. Viruses. 2021;13:146. doi: 10.3390/v13020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fratev F. N501Y and K417N mutations in the spike protein of SARS-CoV-2 alter the interactions with both hACE2 and human-derived antibody: a free energy of perturbation retrospective study. J Chem Inf Model. 2021;61:6079–6084. doi: 10.1021/acs.jcim.1c01242. [DOI] [PubMed] [Google Scholar]
- 34.Jangra S, Ye C, Rathnasinghe R, et al. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. 2021;2:e283–e284. doi: 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andreano E, Piccini G, Licastro D, Casalino L, Johnson NV, Paciello I, Dal Monego S, Pantano E, Manganaro N, Manenti A, Manna R, Casa E, Hyseni I, Benincasa L, Montomoli E, Amaro RE, McLellan JS, Rappuoli R. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. biorxiv. 2020;28:424451. doi: 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baral P, Bhattarai N, Hossen ML, Stebliankin V, Gerstman BS, Narasimhan G, Chapagain PP. Mutation-induced changes in the receptor-binding interface of the SARS-CoV-2 delta variant B. 1.617. 2 and implications for immune evasion. Biochem Biophysic Res Commun. 2021;574:14–19. doi: 10.1016/j.bbrc.2021.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshi M, Puvar A, Kumar D, Ansari A, Pandya M, Raval J, Patel Z, Trivedi P, Gandhi M, Pandya L, Patel K, Savaliya N, Bagatharia S, Kumar S, Joshi C. Genomic variations in SARS-CoV-2 genomes from gujarat: underlying role of variants in disease epidemiology. Front Genet. 2021;12:586569. doi: 10.3389/fgene.2021.586569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghanchi NK, Nasir A, Masood KI, Abidi SH, Mahmood SF, Kanji A, Razzak S, Khan W, Shahid S, Yameen M, Raza A, Ashraf J, Ansar Z, Dharejo MB, Islam N, Hasan Z, Hasan R. Higher entropy observed in SARS-CoV-2 genomes from the first COVID-19 wave in Pakistan. PLoS One. 2021;16:e0256451. doi: 10.1371/journal.pone.0256451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duchene S, Featherstone L, Haritopoulou-Sinanidou M, Rambaut A, Lemey P, Baele G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020;6:veaa061. doi: 10.1093/ve/veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Wang W, Zhao X, Zai J, Zhao Q, Li Y, Chaillon A. Transmission dynamics and evolutionary history of 2019-nCoV. J Med Virol. 2020;92:501–511. doi: 10.1002/jmv.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nie Q, Li X, Chen W, Liu D, Chen Y, Li H, Li D, Tian M, Tan W, Zai J. Phylogenetic and phylodynamic analyses of SARS-CoV-2. Virus Res. 2020;287:198098. doi: 10.1016/j.virusres.2020.198098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ko K, Nagashima SEB, Ouoba S, Akita T, Sugiyama A, Ohisa M, Sakaguchi T, Tahara H, Ohge H, Ohdan H, Kubo T, Kishita E, Kuwabara M, Takahashi K, Tanaka J. Molecular characterization and the mutation pattern of SARS-CoV-2 during first and second wave outbreaks in Hiroshima, Japan. PLoS One. 2021;16:e0246383. doi: 10.1371/journal.pone.0246383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the GenBank database (https://www.ncbi.nlm.gov/genome) under accession numbers OK083891-OK084640, OM984745-OM984850, and OM996047-OM996083.