FIGURE 6.
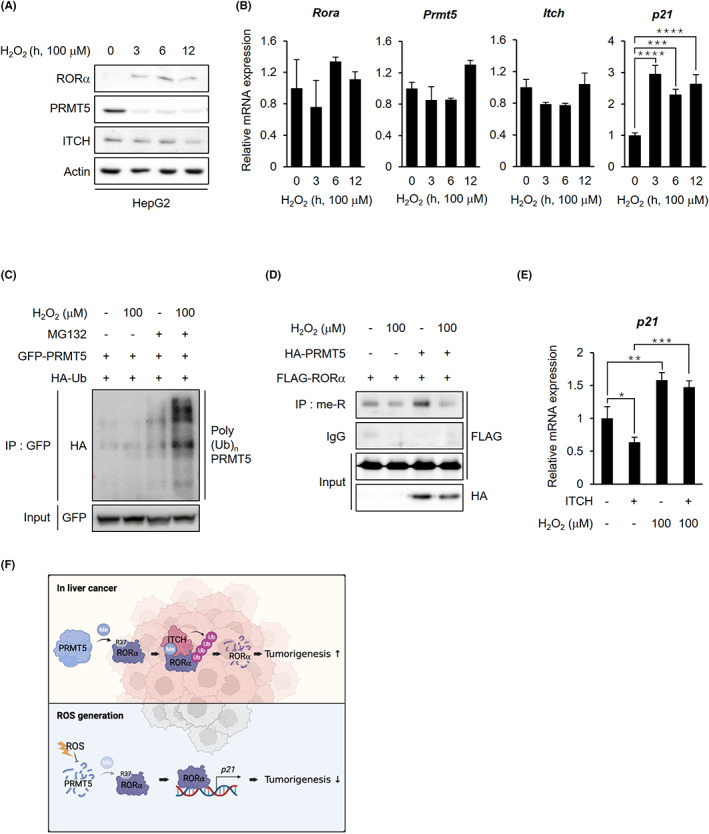
ROS facilitates RORα stabilization through PRMT5 inhibition in liver cancer cells. A, Endogenous protein levels of RORα, PRMT5, and ITCH in HepG2 cells after H2O2 treatment. Cells were treated with 100 μM H2O2 for the indicated lengths of time. β‐actin was used as a loading control. B, qRT‐PCR analysis for relative Rora, Prmt5, Itch, and p21 mRNA levels in HepG2 cells treated with 100 μM H2O2 for 0, 3, 6, and 12 h. The mRNA levels were normalized to GAPDH expression. C, Ubiquitination assay of GFP‐PRMT5 in 293 T cells cotransfected with HA‐Ub after H2O2 treatment. Cells were incubated with or without 100 μM H2O2 and 5 μg/ml proteasomal inhibitor MG132 for 6 h. D, Arginine methylated FLAG‐RORα levels in 293 T cells after cotransfection with HA‐mock or PRMT5 WT. Cells were incubated with or without 100 μM H2O2 for 6 h. E, qRT‐PCR analysis for relative p21 mRNA levels in HepG2 cells after transfection with FLAG‐mock or ITCH. Cells were treated with or without 100 μM H2O2 for 6 h. The mRNA levels were normalized to HPRT expression. F, The schematic diagram of the mechanism of arginine methylation‐mediated RORα degradation. In liver cancer, PRMT5‐catalyzed arginine methylation of RORα induces RORα ubiquitination by facilitating RORα and E3 ligase ITCH interactions. When the cells were exposed to excessive ROS, the cells inhibited PRMT5 from dissociating ITCH from RORα, resulting in RORα stabilization. Statistical analysis was performed using one‐way ANOVA and two‐way ANOVA with Tukey's post hoc tests. *p < 0.05. Data are expressed as mean ± SEM
