Figure 3.
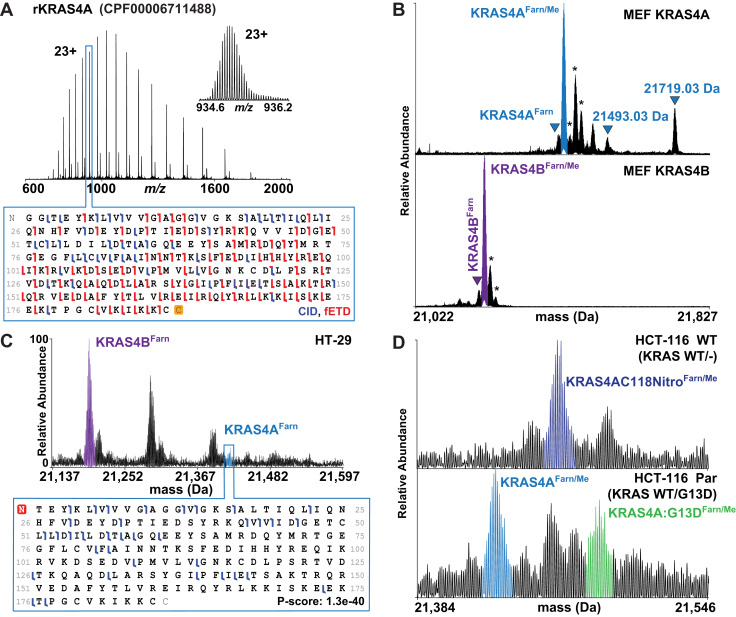
KRAS4AProteoforms.A, recombinant KRAS4A charge state distribution and graphical fragment map from a 21 T FT–ICR mass spectrometer. B, proteoform landscapes in MEF KRAS4A WT and MEF KRAS4B WT cell lines from a 21 T FT–ICR mass spectrometer. Peaks in the KRAS4A landscape match the masses for KRAS4A with phosphorylation (21,493.03 Da) as well as C185 geranylgeranylation with C181 palmitoylation (21,719.03 Da). C, KRAS4B versus KRAS4A proteoform relative abundances in the HT-29 cell line (top) and corresponding KRAS4A fragment ion map (bottom). D, allele-specific KRAS4A proteoforms shown in HCT-116 parental and WT isogenic cell lines. Asterisks denote oxidation. FT–ICR, Fourier transform ion cyclotron resonance; MEF, mouse embryonic fibroblast.