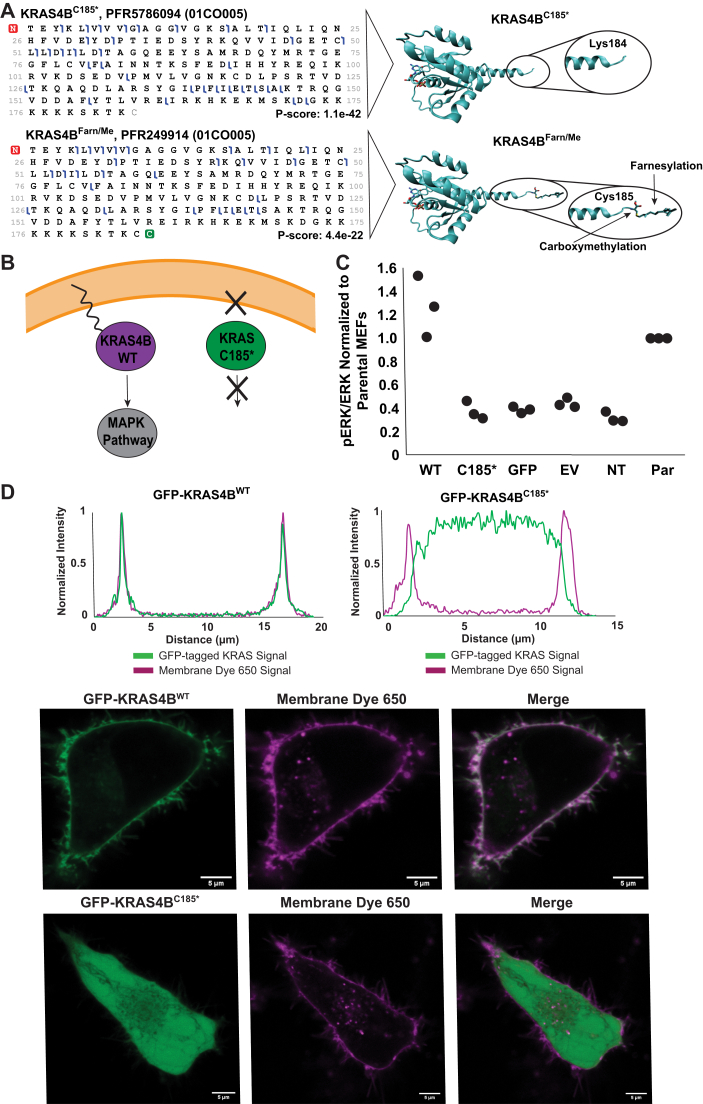
Figure 5.
Novel KRAS4B Truncation.A, graphical fragment maps from TDMS of KRAS4BFarn/Me and KRAS4BC185∗ proteoforms and their corresponding structural models (Protein Data Bank ID: 5TAR). B, scheme depicting hypothesized differences in membrane association and cell signaling between KRAS4BFarn/Me and KRAS4BC185∗. C, scatter plot showing pERK/ERK levels in MEFs transfected with different plasmids (WT = GFP-KRAS4BWT; no Cys185 = GFP-KRAS4BC185∗; GFP = GFP-only vector; EV = empty vector; NT = no transfection control) normalized to pERK/ERK levels in parental (PAR) MEFs (no 4OHT, no transfection). Densitometry measurements were performed by Fiji ImageJ (73). All three replicates are displayed. D, intensity traces of GFP-KRAS and Membrane Dye 650 signal versus distance across a cell as determined by Fiji ImageJ (micrometer) (top) and live-cell images of HeLa cells expressing KRAS4BFarn/Me or KRAS4BC185∗ plasmids (bottom) (bar represents 5 μm). 4OHT, 4-hydroxytamoxifen; MEF, mouse embryonic fibroblast; TDMS, top–down mass spectrometry.