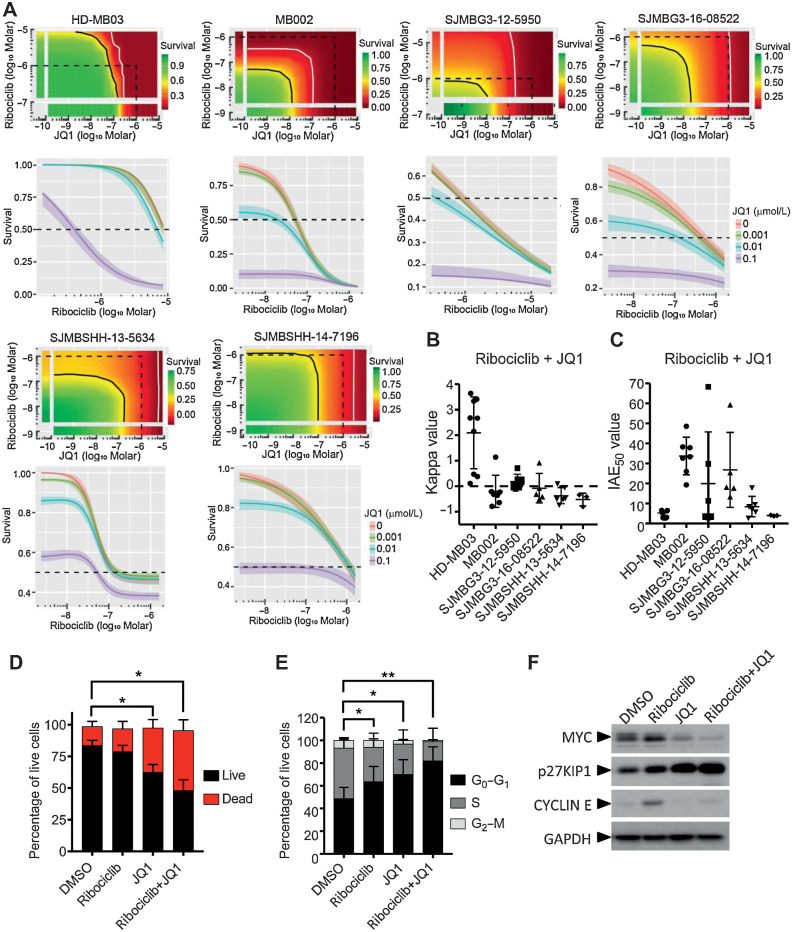
Figure 3.
In vitro combination of ribociclib and JQ1. A–C, In 384-well plate format, HD-MB03, MB002, G3MB, and SHHMB PDXs treated with ribociclib and JQ1 in dose–responses for 7 days. Growth inhibition evaluated with CellTiter Glo. Bivariate Response to Additive Interacting Doses (BRAID) response surface model assessing the effect and efficacy of the combination. A, Representative response surface model (top) and JQ1 potentiation of ribociclib dose–response (bottom) shown for each model. The red and green color scales indicate low and high proliferation, respectively, relative to vehicle-treated cells. The dotted line encompasses the region in the response surface where each drug is ≤1 μmol/L and shown for reference. The black and white lines are the drug combination isoboles representing 50% and 90% inhibition of cell proliferation, respectively (top). Endpoints of these lines are driven by the potency of each drug as single agent. These lines will curve inward toward the origin for synergistic combinations and bow outward away from the origin for antagonistic combinations. B, Kappa values (n = 3–9), type of interaction between the two combined drugs. When kappa is close to zero, the combination is additive, whereas positive and negative values imply synergism and antagonism, respectively. C, IAE50 values (n = 3–9), measure of the efficacy at achieving 50% growth inhibition. BRAID data are available in Supplementary Table S1. D–F, HD-MB03 cells treated with DMSO, 5 μmol/L of ribociclib, 0.3 μmol/L of JQ1 or the combination for 72 hours. D, Dead and apoptotic cells stained with Annexin V-APC reagent and (E) DNA content analyzed on permeabilized nuclei using propidium iodide by flow cytometry. Statistical analyses performed on the percentage of AnnexinV positive cells (dead; D) and on the percentage of cells in G0–G1 in the cell cycle (E; Mann and Whitney test, n = 4 and n = 6, respectively). F, MYC, CYCLIN E, p27KIP1, and GAPDH expression analyzed by immunoblotting (n = 3).