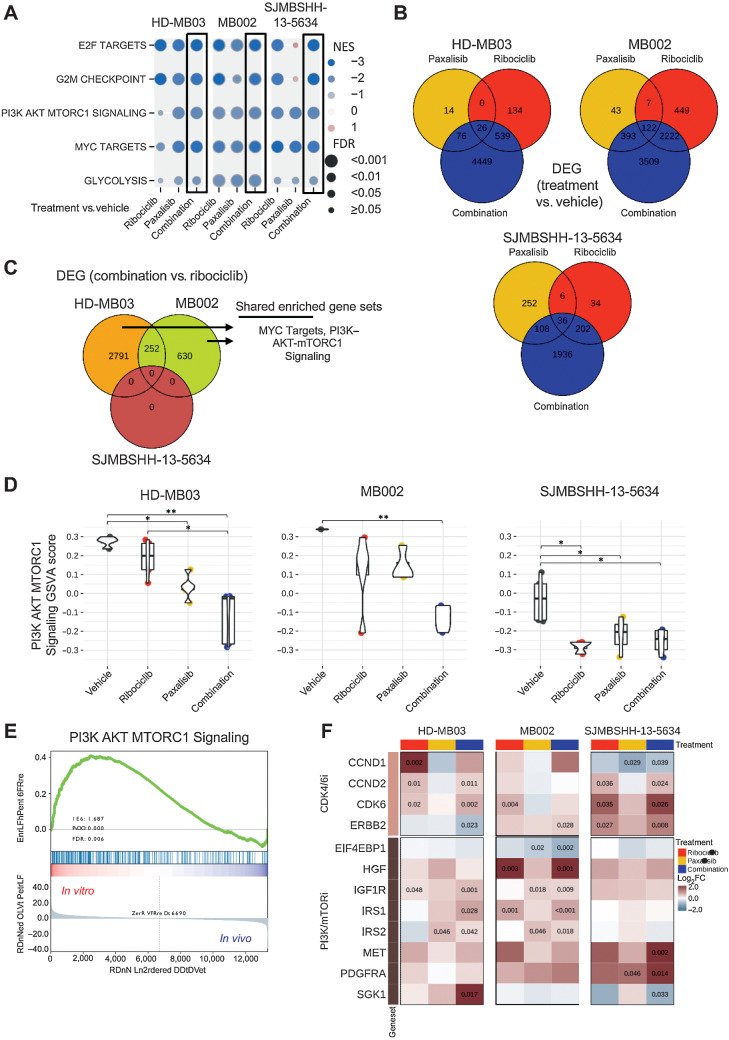
Figure 7.
Gene expression analysis of tumors treated with ribociclib and paxalisib. RNAs from HD-MB03, MB002, and SJMBSHH-13–5634 tumors, from the preclinical studies presented on Fig. 6, were sequenced. Gene set enrichment analysis (GSEA) and differential expressed gene (DEG) analysis performed between tumors treated with ribociclib, paxalisib or combination versus vehicle. A, Plot showing the Normalized Enrichment Score (NES, circles in color) and FDR (circles size) of the gene sets found to be significantly depleted in each of the three medulloblastoma models when comparing combination versus vehicle-treated tumors (black boxes). Other comparisons include ribociclib or paxalisib versus vehicle-treated tumors. GSEA data are presented in Supplementary Table S4. B, Venn diagram of DEGs between each treatment (ribociclib-only, paxalisib-only, and the combination) and vehicle for each model (FDR < 0.05). C, Venn diagram of DEGs between combination and ribociclib-treated tumors in each model. Enriched gene set term analysis of HD-MB02 and MB002 models DEGs, run separately, showed two shared gene sets, including MYC Targets and PI3K–AKT–MTORC1 signaling. Data are presented in Supplementary Tables S5 and S6. D, Multiple pairwise comparison of PI3K–AKT–MTORC1 signaling pathway between treatments for each medulloblastoma model. Gene set variation scores of PI3K–AKT–MTORC1 pathway genes calculated for each sample and compared with a Kruskal–Wallis test, followed by a post hoc Dunn test. Only significant FDR adjusted P comparisons are shown (*, P < 0.05; **, P < 0.01). E, GSEA of PI3K–AKT–MTORC1 signaling with HD-MB03 cell line (in vitro) and HD-MB03 vehicle treated tumors (in vivo). F, Heatmap with log2 fold change (log2FC) between ribociclib, paxalisib or combination versus the vehicle group for genes known to be involved in resistance to CDK4/6 and PI3K/mTOR inhibitors. Adjusted P values are indicated for significant DEGs.