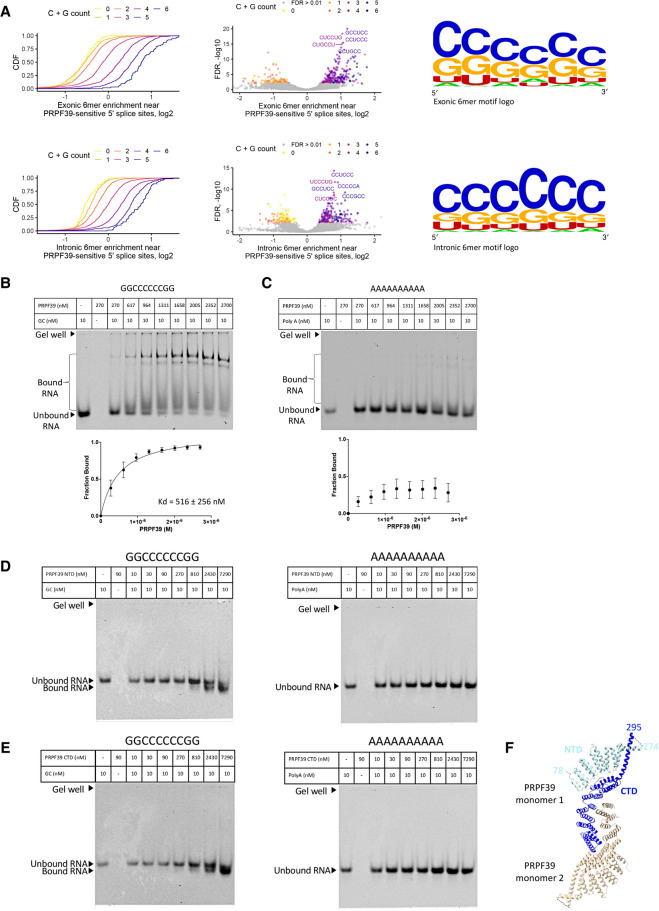
FIGURE 3.
PRPF39 interacts with RNA. (A) Intronic and exonic 6mer enrichment near PRPF39-sensitive 5′ splice sites demonstrated using CDF (left), volcano plot (middle), and logo (right, derived from WebLogo [Schneider and Stephens 1990; Crooks et al. 2004]). (B) EMSA of full-length PRPF39 with a GC-rich oligo (GGCCCCCCGG) demonstrates that PRPF39 binds the GC oligo with a Kd of 516 ± 256 nM. Identity of the protein bound RNA was analyzed using western blot analyses (Supplemental Fig. S1). (C) EMSA of full-length PRPF39 with a 10-mer poly(A) control oligo. (D) EMSA of PRPF39 NTD with the GC and poly(A) oligo. (E) EMSA of PRPF39 CTD with the GC and poly(A) oligo. Note that the migration of a molecule in native gel is determined by the combined effect of its size, charge, and shape, and the substantial negative charge of NTD and CTD (pI of 4.62 and 6.15, respectively) likely have led to a faster migrating band despite the increased mass when bound to the RNA. (F) The dimeric human PRPF39 homologous model based on the murine PRPF39 structure (PDB: 6G70) showing the boundary of the NTD (cyan) and CTD (blue) in one monomer. Dashed line indicates the disordered region separating the NTD and CTD with the flanking residue numbers labeled.