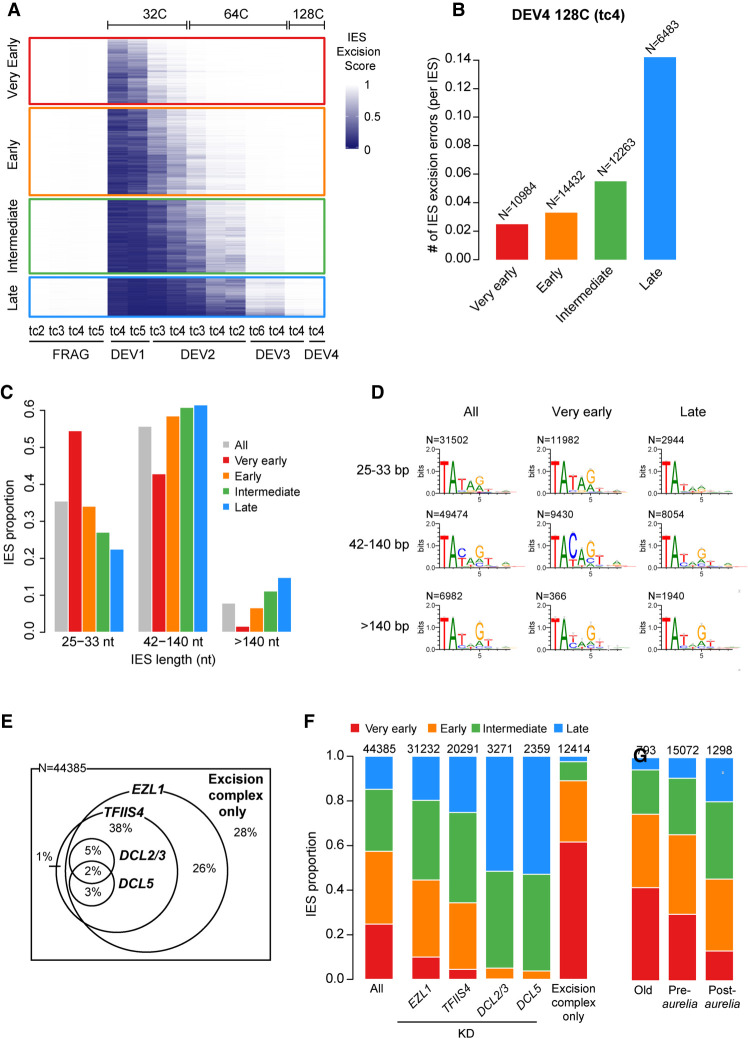
Figure 4.
Excision timing defines IES classes with different characteristics. (A) Heatmap of ESs for all IESs. IESs are sorted by hierarchical clustering, with each row corresponding to one IES. The ES is encoded from zero (dark blue; no excision) to one (white; complete excision). IESs are separated in four classes according to their excision profile by k-means clustering of their ES (very early: N = 10,994; early: N = 14,490; intermediate: N = 12,353; late: N = 6548). (B) Abundance of IES excision errors in the four excision profile groups counted in the DEV4 128C (tc4) sample. In this analysis, we focused on error types that would be the least impacted by IES length (external, overlap, and partial external) (see Supplemental Fig. S7A). The number of IESs in each excision profile group is indicated above the bars. (C) IES fraction for IES length categories in the four excision profile groups compared with all IESs. (D) Sequence logos of the eight bases at IES ends for all IESs and IESs belonging to the very early and late clusters. IESs are grouped in three length categories as described in C. (E) Venn diagram showing how the 44,385 reference IESs are distributed according to their sensitivity to EZL1, TFIIS4, DCL2/3, and DCL5 RNAi with regard to excision. The group “excision complex only” represents IESs that do not depend on any of these factors but do depend upon Pgm. The Venn diagram has been simplified to display only overlaps representing >1% of the total number of IESs. (F) IES proportions in the four groups of excision profiles for the data sets defined in E. The numbers above the barplots indicate the number of IESs in each data set. “All” is the random expectation for all IESs. (G) IES proportions in the four groups of excision profiles relative to the age of IES insertion during evolution of the Paramecium lineage. (Old) Insertion predating the divergence between P. caudatum and the P. aurelia clade; (pre-aurelia) insertion before the radiation of the P. aurelia complex; (post-aurelia) insertion after the radiation of the P. aurelia complex (Sellis et al. 2021).