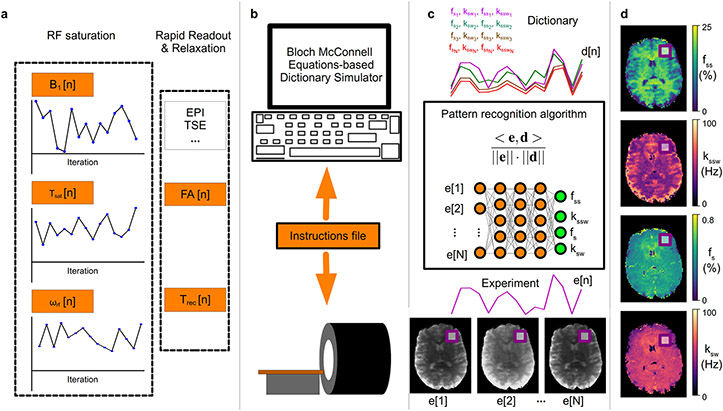
Figure 1. General pipeline of a semisolid MT/CEST MRF experiment.
a. Initially, a pseudo-random imaging protocol is designed, where at least one acquisition parameter is being varied, to produce a set of N images. Importantly, a pre-saturation block needs to be implemented, where at least the saturation pulse power (B1), duration (Tsat), or frequency offset (ωrf) should vary, for sufficient encoding of the chemical exchange parameters. The protocol typically includes a rapid readout, e.g., using EPI or turbo spin echo, with either a fixed or varied flip angle (FA) and recovery time (Trec). b. The designed protocol is then loaded into a computerized Bloch-McConnell equations-based signal simulator, which produces the signal trajectories expected for a large number of tissue parameter combinations. The same CEST-MRF acquisition protocol is fed as an instruction file to the MRI scanner, allowing the acquisition of N molecular information encoding images, where each pixel series comprises an experimentally acquired trajectory (e[1] – e[N]). c. Each trajectory (e[n]) is then compared to all dictionary entries (d[n]), via a pattern recognition algorithm (such as the dot-product metric) for the determination of the best match. Importantly, this step can be accelerated and improved using a deep neural-network. d. Finally, simultaneous pixel-wise quantification of the proton exchange rates (k), and volume fractions (f) for a single or several metabolite/protein/lipid pools of interest can be made, based on the neural network output, or the best-matched dictionary entry.