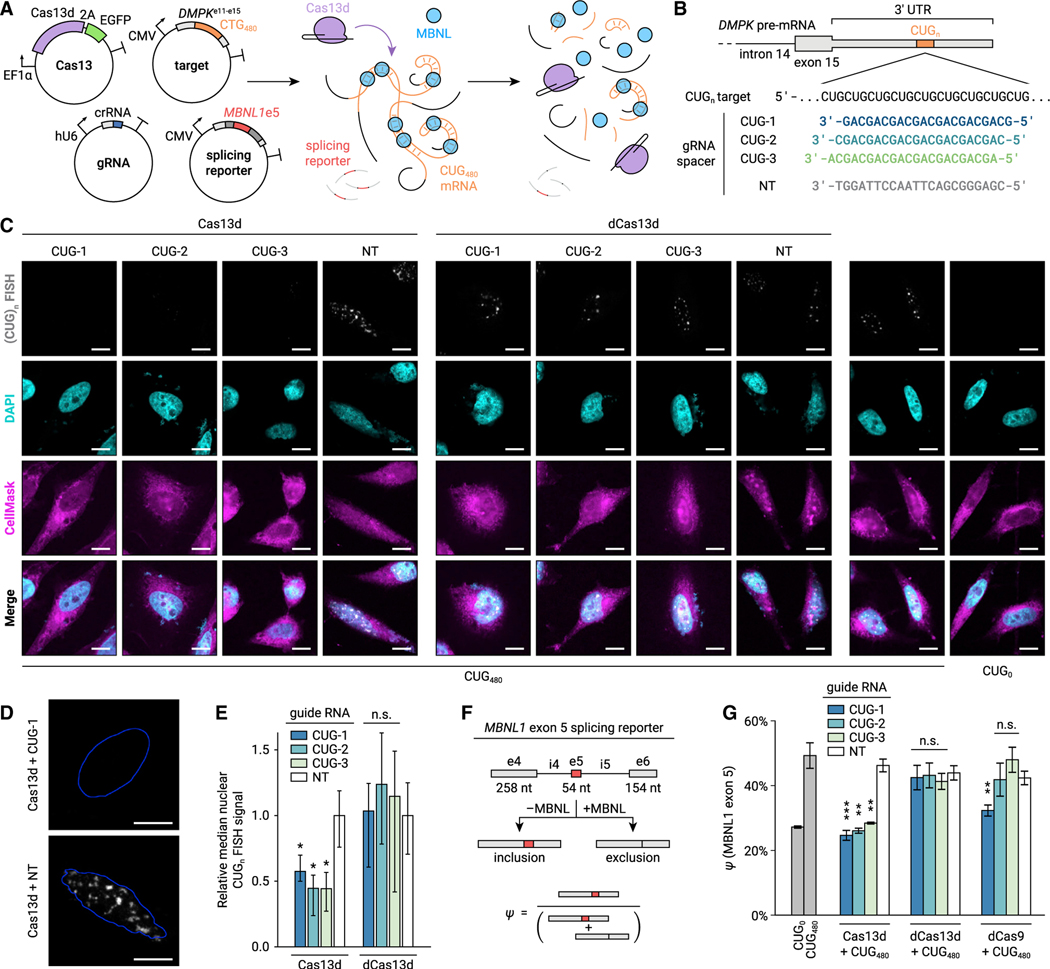
Figure 1. Cas13d reduces accumulation of CUGn repeat RNA and rescues MBNL-dependent splicing in a HeLa cell-culture model of DM1.
(A) Diagram of cotransfection experiments to test Cas13d on-target activity and MBNL-dependent splicing. Plasmids encoding Cas13d and EGFP, gRNA, CUG480 target RNA, and an MBNL1 exon 5 splicing minigene are expressed in HeLa cells prior to CUGn RNA FISH and RT-PCR.
(B) gRNAs tested, including three CUG-targeting gRNAs and a non-targeting (NT) control.
(C) Representative FISH images for CUGn RNA (grayscale) in targeting and NT conditions. Cells stained with DAPI (nuclei, cyan) and CellMask (cells, magenta) for segmentation. Scale bars, 10 μm.
(D) Magnified images of CUGn RNA FISH (grayscale) from (C) in targeting and NT conditions. Outlines of nuclei (blue) computed by automatic thresholding. Scale bars, 10 μm.
(E) Median CUGn FISH signal per nucleus, relative to NT. Error bars show 95% confidence interval (CI), estimated by bootstrapping. n > 7 nuclei. *p < 0.05, non-overlapping CI. n.s., not significant (p > 0.05).
(F) Diagram of splicing minigene assay. Sequestration of MBNL by CUGn RNA increases MBNL1 exon 5 inclusion ratio (ψ). CUG-targeting Cas13d aims to rescue ψ to unperturbed levels.
(G) MBNL1 exon 5 ψ for targeting and NT conditions, measured by RT-PCR. Transcriptional inhibition by deactivated Cas9 (dCas9) with matching spacers is also shown. Error bars show standard deviation (SD). n = 3. *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed Student’s t test.