Figure 3. Urea accumulation causes mitochondrial dysfunction.
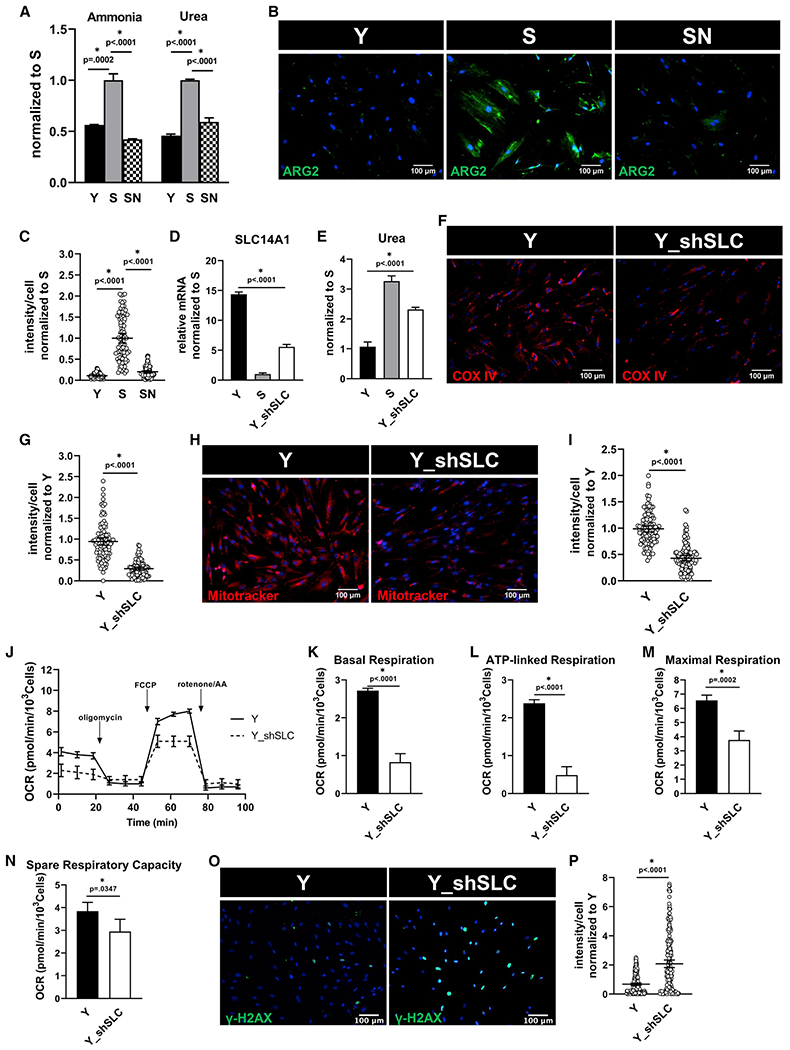
(A) Intracellular ammonia and urea concentration measured in Y, S, and SN cells. Data normalized to S and represented as mean ± SD.
(B) Immunostaining for urea cycle enzyme, arginase-2 (ARG-2); scale bar represents 100 μm.
(C) Quantification of ARG-2 intensity per cell normalized to S; data shown as mean ± 95% CI for >100 cells.
(D) Quantification of gene expression of SLC14A1 via quantitative RT-PCR normalized to S and internally normalized to RPL32 cycle number. Y_shSLC denotes Y cells with SLC14A1 knockdown.
(E) Intracellular urea concentration measured in Y, S, and Y_shSLC cells, normalized to Y cells.
(F) Immunostaining for COX IV; scale bar represents 100 μm.
(G) Quantification of COX IV intensity per cell normalized to Y; data shown as mean ± 95% CI for >100 cells.
(H) Mitotracker Red live stain images; scale bar represents 100 μm.
(I) Quantification of Mitotracker Red intensity per cell; data shown as mean ± 95% CI for >100 cells.
(J) Measurement of oxygen consumption (OCR) using Seahorse extracellular flux analyzer.
(K–N) Basal respiration (K), ATP-linked respiration (L), maximal respiration (M), and spare respiratory capacity (N) rates calculated from the OCR data in (L).
(O) Immunostaining for phosphorylated form of histone H2AX (γ-H2AX), a measure of DNA damage; scale bar represents 100 μm.
(P) Quantification of γ-H2AX intensity per cell, normalized to Y; data shown as mean ± 95% CI for >100 cells.
