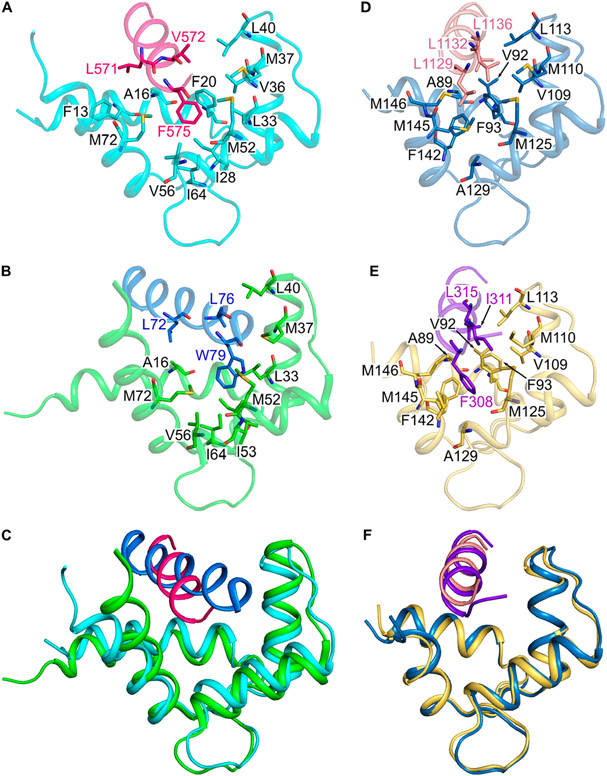
Figure 6.
Comparison of CaM/CaM1 vs CaM/CNGA2 and CaM/CaM2 vs CaM/CK. Main chain structures of (A) CaM/CaM1 (PDB ID: 8DGK), (B) CaM/CNGA2 (PDB ID: 1SY9), (D) CaM/CaM2 (PDB ID: 8DGH), and (E) CaM/CK (PDB ID: 7BF2). Main chain structures of CaM/CaM1 overlaid with CaM/CNGA2 (C) and CaM/CaM2 overlaid with CaM/CK (F). The CaM N-lobe (cyan) is bound to CaM1 (magenta helix) in panels A and C; the CaM N-lobe (green) is bound to CNGA2 (blue helix) in panels B and C; the CaM C-lobe (dark cyan) is bound to CaM2 (light red helix) in panels D and F; the CaM C-lobe (yellow) is bound to the peptide fragment of the creatine kinase (CK, purple helix) in panels E and F.