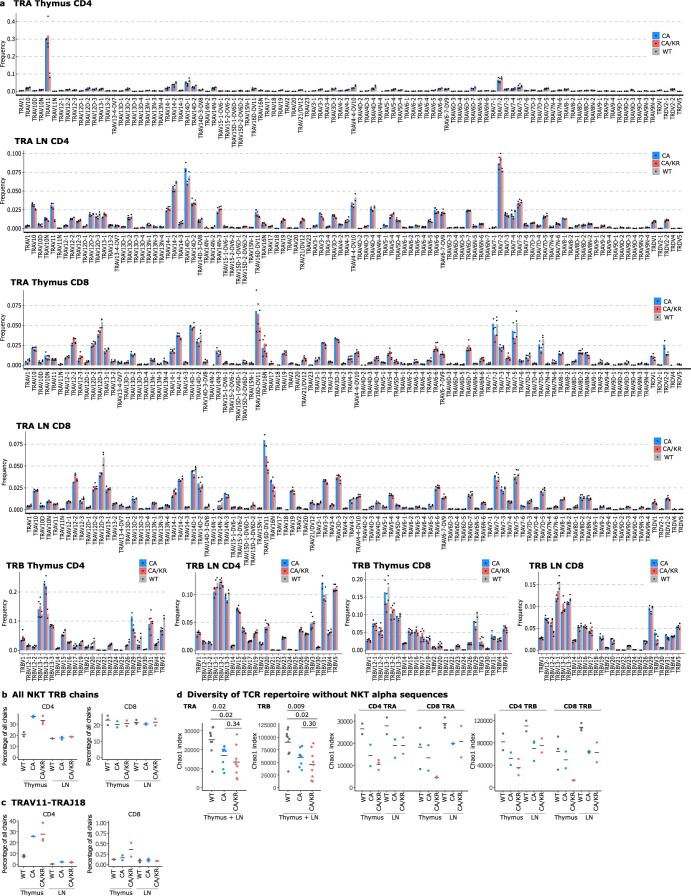
Extended Data Fig. 6. Gene segment usage analysis of TCR repertoires in Lck variant mice.
(a-d) Analysis of TCR repertoires in CD4+ and CD8+ LN T cells, and mature SP4 and SP8 thymocytes from LckWT/WT (gray), LckCA/CA (blue) and LckCA/KR (red) mice. The information about the sample size are in Supplementary Table 6. Individual mice and means are shown. Related to Fig. 2e-g. (a) The usage of TRAV and TRBV gene segments in the indicated mice. Each bar represents the average frequency of the gene segment among all CDR3 sequences in a particular group of samples. (b) A total percentage of typical TCRβ (TRB) gene segments used by NKT cells (TRBV1,TRBV13-2, and TRBV9) in the indicated samples from the indicated mice. (c) Percentage of TCRs containing TRAV11-TRAJ18 gene segments typically used by NKT cells in the indicated cells from the indicated mice. (d) Diversity of the repertoire of the TCRα (TRA) and TCRβ (TRB) CDR3 amino acid sequences in the indicated cells from the indicated mice calculated using the Chao1 richness estimator. CD4 and CD8 T-cell CDR3 amino acid sequences were analyzed together (left) or separately (right). Invariant NKT TRAV11-TRAJ18 CDR3 amino acid sequences were removed before this analysis. The statistical significance was calculated using a Kruskal-Wallis test with multiple comparison adjustment using the Holm method.