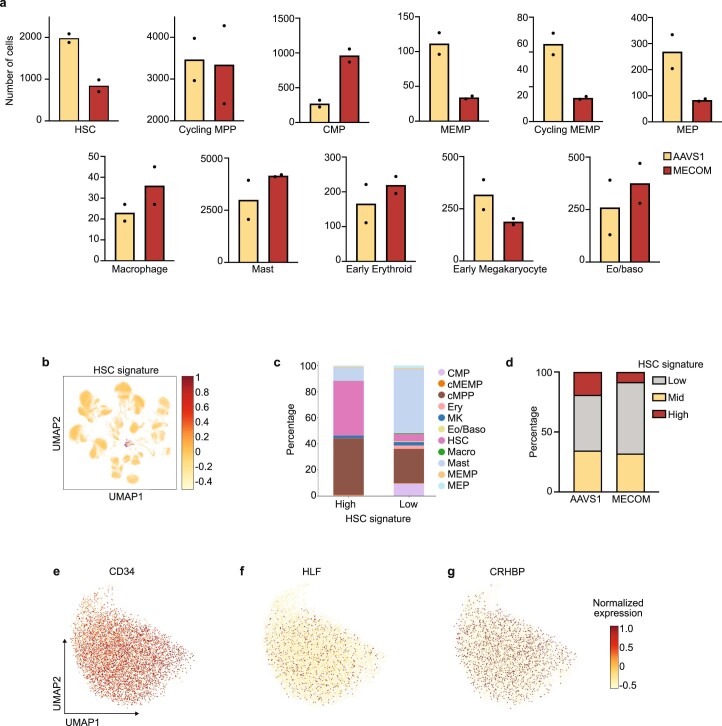
Extended Data Fig. 2. Single-cell RNA sequencing of LT-HSCs after MECOM editing.
(a) Bar graphs showing the number of cells in each of the 11 hematopoietic cell clusters identified by single cell RNA sequencing of CD34+CD45RA−CD90+ HSCs following AAVS1 (yellow) or MECOM (red) editing. Mean is plotted and each of two biological replicates is shown. Total number of cells profiled in each group: AAVS1 – 19,375, MECOM – 19,821. (b) Uniform manifold approximation and projection (UMAP) plot of 263,828 single cells from human umbilical cord blood, colored according to HSC signature (CD34, HLF, CRHBP). (c) Transcriptional identities of cells stratified by HSC signature score. HSC signature score was calculated for CD34+CD45RA−CD90+ HSCs from Fig. 2d. Cells were grouped into high HSC signature score (>0.5), mid HSC signature score (>0 and <0.5), and low HSC signature score (<0) clusters, and cell identities were determined by transcriptional signatures using Celltypist41. Cells with a high HSC signature score were enriched for HSCs and cMPPs. Abbreviations of cell types defined in Fig. 2a. (d) Stacked bar graph showing AAVS1 or MECOM edited CD34+CD45RA−CD90+ HSCs stratified by expression of HSC signature score as defined in Extended Data Fig. 2c. MECOM editing leads to a depletion of cells with high HSC signature score. (e-g) UMAP plots of the normalized expression of CD34 (e), HLF (f), and CRHBP (g) in phenotypic LT-HSCs. The combined expression of these three genes defines the HSC signature in Fig. 2d, e and Extended Data Fig. 2c, d.