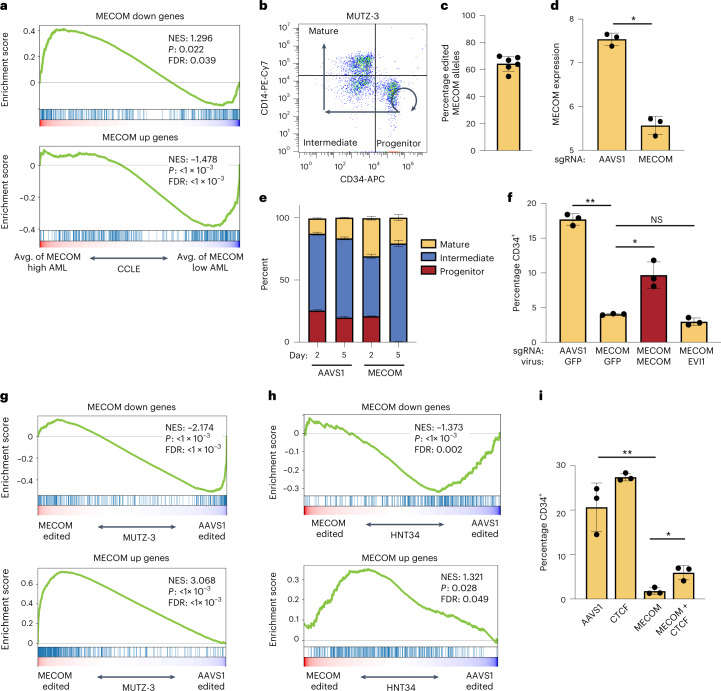
Fig. 8. The MECOM gene regulatory network is indispensable in AML.
a, GSEA of MECOM down genes and MECOM up genes in CCLE AML cell lines. AML cell lines were stratified by MECOM expression as in Extended Data Fig. 8a. MECOM-high AMLs show enrichment of MECOM down genes and depletion of MECOM up genes compared to MECOM-low AMLs. The K–S test was used to determine the significance of GSEA. b, FACS plot showing the immunophenotype of MUTZ-3 cells. CD34+CD14− progenitors can self-renew (curved arrow) and undergo differentiation (straight arrows) into CD34−CD14− intermediate promonocytes and ultimately CD34−CD14+ mature monocytes. c, MECOM editing in MUTZ-3 AML cells. Cells were collected on day 3 after nucleofection of CRISPR ribonucleoprotein (RNP) and the percent of modified alleles was determined by Sanger sequencing and ICE analysis; n = 6 biologically independent samples. Mean is plotted and error bar shows s.e.m. d, MECOM expression (log2 RPKM) in CD34+ MUTZ-3 cells. MECOM editing causes significant reduction in expression; n = 3 per group. Mean is plotted and error bars show s.e.m. Two-sided Student’s t-test was used. *P = 2 × 10−4. e, Myelomonocytic differentiation analysis of MUTZ-3 cells after CRISPR editing. Percent of cells within each subpopulation was measured by flow cytometry on days 2 and 5 after editing. n = 3 per group. Mean is plotted and error bars show s.e.m. f, Percentage of MUTZ-3 cells in CD34+CD14− progenitor population after MECOM editing and viral rescue as determined by flow cytometry; n = 3 per group. Mean is plotted and error bars show s.e.m. Two-sided Student’s t-test was used. *P = 3.6 × 10−2, **P = 1.5 × 10−3. g,h, GSEA of MECOM network genes in MUTZ-3 cells (g) and HNT34 cells (h) after MECOM editing. MECOM perturbation in both AML cell lines results in enrichment of MECOM down genes and depletion of MECOM up genes. The K–S test was used to determine the significance of GSEA. i, Bar graphs of the rescue of CD34+ by dual MECOM and CTCF perturbation. MUTZ-3 AML cells underwent CRISPR editing with the sgRNA guides depicted on the x axis. Percent CD34+ cells were determined by FACS on day 4; n = 3 per group. Mean is plotted and error bars show s.e.m. Two-sided Student’s t-test was used. *P = 1.4 × 10−2, **P = 3.9 × 10−3.