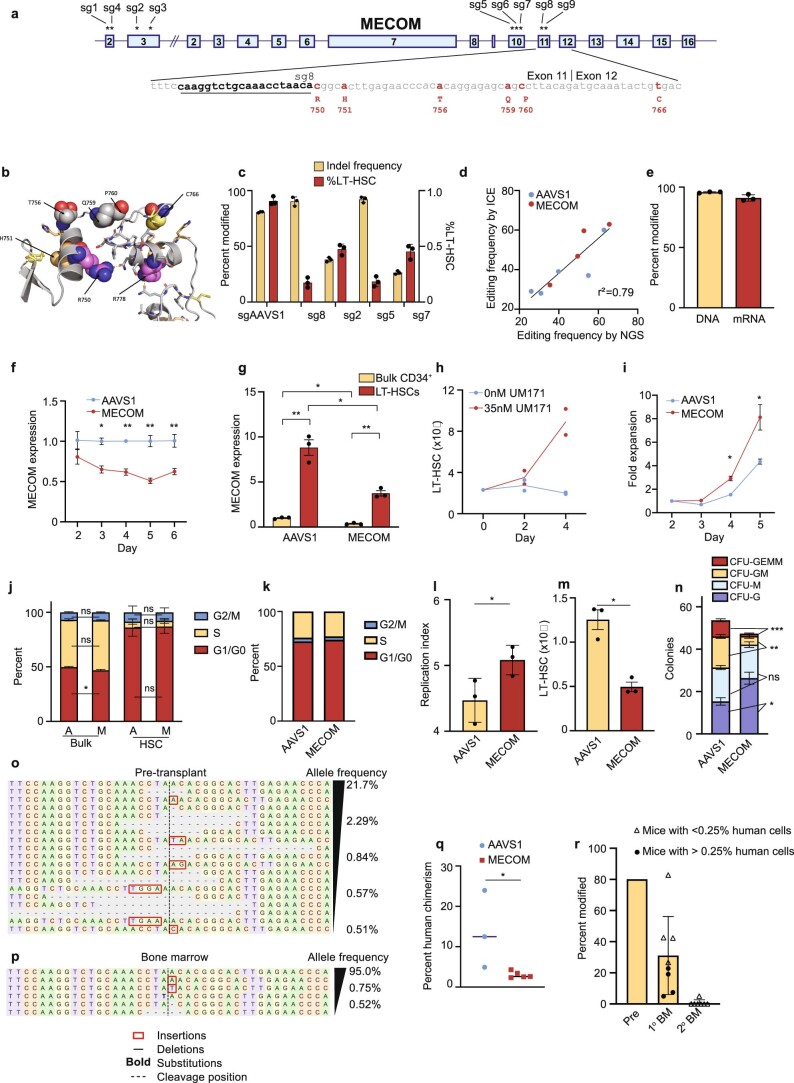
Extended Data Fig. 1. Modeling MECOM haploinsufficiency in human CD34+ HSPCs.
(a) Schematic of the MECOM locus annotated with the location of sgRNAs (sg1-sg9) tested for efficiency of MECOM editing. The binding site of sg8 (underlined) which is used in subsequent studies is shown, and clinical mutations annotated with amino acid number that have been described in MECOM haploinsufficient bone marrow failure (red) are indicated. (b) Predicted partial protein structure of the MECOM zinc finger domain with mutated residues shown as spheres. These mutations are expected to disrupt the structure of the zinc finger, either through abrogation of Zn coordination (H751, C766) or tethering between the ZnF (R750, R778). (c) Percent modified alleles (left y-axis) and percent LT-HSCs of total live cells (right y-axis) after CRISPR editing of primary human CD34+ HSPCs. Editing efficiency was detected at 72 hours after RNP delivery of Cas9 and sgRNA by nucleofection and percent of live cells that remained in the LT-HSC gate was evaluated on day 6. LT-HSCs are defined by the following immunophenotype: CD34+CD45RA−CD90+CD133+EPCR+ITGA3+. sg2, sg5, sg7, sg8 are sgRNAs targeting MECOM as described in Extended Data Fig. 1a. n = 3 biologically independent samples. Mean is plotted and error bars show s.e.m. (d) Comparison of Sanger sequencing followed by ICE analysis and Next Generation Sequencing (NGS) for the detection of CRISPR edits. AAVS1 (blue) and MECOM (red) edited samples were analyzed by ICE and NGS in parallel. (e) MECOM editing in human CD34+ HSPCs after RNP delivery by nucleofection. Editing frequency was detected at 48 hours by Sanger sequencing of genomic DNA. Transcription of edited MECOM alleles was determined by cDNA synthesis followed by Sanger sequencing of RNA from bulk HSPCs at 48 hours. n = 3 biologically independent samples. Mean is plotted and error bars show s.e.m. (f) MECOM expression following CRISPR editing. MECOM expression (normalized to GAPDH) in bulk HSPCs was detected by qRT-PCR (n = 3 AAVS1, n = 9 MECOM; three biologically independent experiments) and was normalized to expression in the AAVS1-edited sample on the same day. Mean is plotted and error bars show s.e.m. Two-sided Student t-test used. *P = 1.7e-3, **P = 2.5e-4. (g) MECOM expression in LT-HSCs. MECOM expression (normalized to GAPDH) was detected by qRT-PCR (n = 3 per group; three biologically independent experiments) in bulk CD34+ HSPCs and in LT-HSCs sorted on day 3 after CRISPR editing. Mean is plotted and error bars show s.e.m. Two-sided Student t-test used. *P = 5.1e-3, **P = 8.3e-4. (h) Expansion of LT-HSCs in culture. HSPCs were cultured in the presence (n = 2) or absence (n = 2) of the HSC self-renewal agonist UM171. Percent of LT-HSCs was determined by FACS as in Fig. 1e and was used to calculate the total LT-HSC number. Cells were supplemented with fresh media every 2 days. (i) Expansion time course of bulk CD34+ HSPCs following CRISPR editing. HSPCs were thawed into HSC media containing 35 nM UM171 and underwent CRISPR editing 24 hours later. Cells were counted daily by trypan blue exclusion starting on day 2 after CRISPR editing and media was added to maintain equal confluency. n = 3 per group. Mean is plotted and error bars show s.e.m. Error bars that are shorter than the size of the symbols have been omitted for clarity. Two-sided Student t-test used. *P = 5e-3. (j) Stacked bar graph of cell cycle status of bulk HSPCs and HSC (HSC: CD34+CD45RA−CD90+CD133+) as determined by Edu incorporation and 7-AAD staining. On day 5 after CRISPR editing, cells were incubated with Edu for 2 hours, then fixed and permeabilized prior to 7-AAD and cell surface staining. AAVS1-edited (A) and MECOM-edited (M) samples, were compared by the proportion of cells in G0/G1 (Edu−/2n DNA content), S (Edu+), or M (Edu−/>2n DNA content) in bulk CD34+ cells or CD34+CD45RA−CD90+ HSCs. n = 3 per group. Mean is plotted and error bars show s.e.m. Two-sided Student t-test used. *P = 8.1e-3. (k) Stacked bar graph of cell cycle status of LT-HSCs as determined by transcriptional signatures of single-cell LT-HSCs. UCB CD34+ underwent CRISPR perturbation of MECOM or AAVS1 and were maintained in HSC media. On day 4 after editing, LT-HSCs were sorted and 10x scRNA sequencing was performed. There was no difference in cell cycle state in LT-HSCs following AAVS1 or MECOM editing. (l) Analysis of cell expansion following CRISPR editing. AAVS1 or MECOM edited HSPCs were labeled with CFSE and successive generations of cell divisions were determined by CFSE signal intensity on day 5 which was used to calculate the replication index, showing the total number of divided cells/cells that underwent at least one division. Mean of three independent experiments is plotted and error bars show s.e.m. Two-sided Student t-test used. *P = 5e-2. (m) Total number of LT-HSCs following MECOM editing. Primary human CD34 + HSPCs underwent CRISPR editing on day 1 after thawing and were cultured in HSC media containing UM171 which was changed every 2 days. On day 6 after editing, the percentage of immunophenotypic LT-HSCs determined by flow cytometry, and the total cell number determined by trypan blue exclusion were used to calculate the total number of LT-HSCs in culture. Mean of three independent experiments is plotted and error bars show s.e.m. Two-sided Student t-test used. *P = 4.7e-3. (n) Stacked bar plots of colony-forming assay comparing MECOM edited HSPCs derived from peripherally mobilized CD34+ cells from healthy adult donors. (n = 6) to AAVS1-edited controls (n = 3). CFU-GEMM, colony-forming unit (CFU) granulocyte erythroid macrophage megakaryocyte; CFU-GM, CFU granulocyte macrophage; CFU-M, CFU macrophage; CFU-G, CFU granulocyte. Mean colony number is plotted and error bars show s.e.m. Two-sided Student t-test used. *P = 3.9e-2, **P = 2.5e-4, ***P = 1.7e-5, ns=not significant. (o-p) NGS of MECOM in human HSPCs following CRISPR editing, prior to xenotransplantation (o), and after harvest from bone marrow at 16 weeks of one representative mouse (p). Sequences present at frequencies >0.5% are displayed. (q) Analysis of bone marrow of mice at week 16 following transplantation of MECOM-edited (n = 5) and AAVS1-edited (n = 3) adult HSPCs. Mean is indicated by black line and each data point represents one mouse. Two-sided Student t-test used. *P = 3.8e-2. (r) Analysis of the MECOM locus of human cells harvested from mice following primary or secondary xenotransplantation. Half of the primary recipient mice (4/8) had human chimerism >0.25% (circles) and the other half had chimerism <0.25% (triangles) but had human MECOM sequences that were detectable by PCR. All of the secondary recipients had human chimerism <0.25% but had human MECOM sequences that were detectable by PCR.