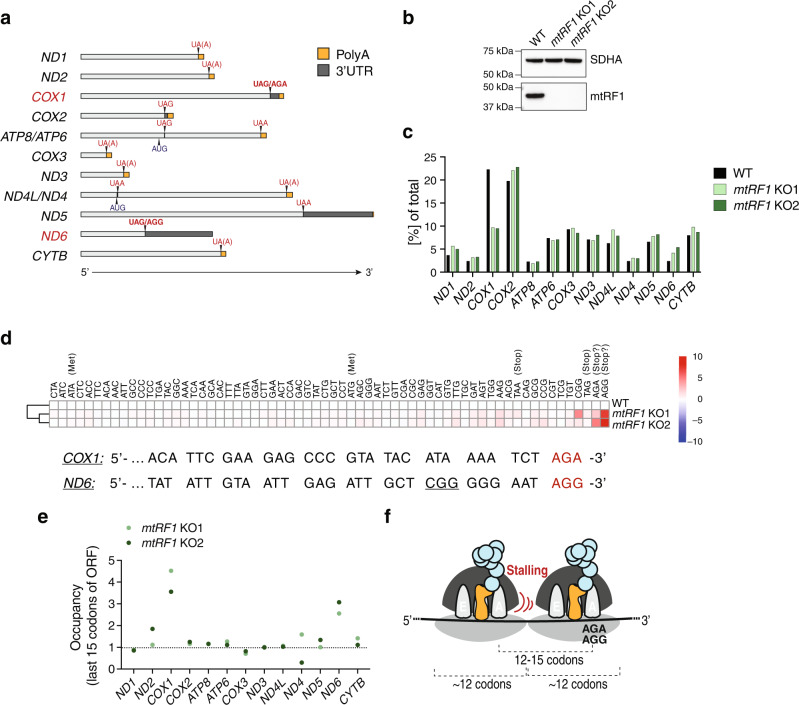
Fig. 1. Mitoribosomes stall at the ends of COX1 and ND6 transcripts in mtRF1 KO cells.
a Schematic representation of human mitochondrial transcripts including polyA tails (orange), 3′ untranslated regions (3′UTRs) (dark gray), stop codons (red), and start codons of bicistronic transcripts (dark blue). Relative lengths of regions are depicted. b Western blot confirming the generation of mtRF1 KO cells. 40 µg mitolysates were loaded. Two different clones are shown (KO1 and KO2). Loading was assessed by SDHA detection. A representative blot of n = 3 independent experiments is depicted. c–e Analysis of ribosome profiling data of WT and mtRF1 KO cells. c Occupancy of mitoribosomes on each transcript relative to the total occupancy on all transcripts. d Upper panel: Codon counts relative to WT cells. Codons are organized according to sequencing coverage (high (left) to low number of reads (right)). Lower panel: Sequences of COX1 and ND6 transcript ends. Of note, codon CGG (underlined) occurs only once in human mitochondrial transcripts. e Occupancy of mitoribosomes on the last 15 codons of each transcript relative to the total occupancy on the respective transcript. Fold change values relative to WT cells are shown. Occupancies on each individual codon are presented in Supplementary Fig. 4. f Schematic representation of stalling events 12–13 codons upstream of stalling on AGA/AGG codons. Ribosome-protected fragments (RPFs) of mitoribosomes usually comprise 35 nucleotides (~12 codons).