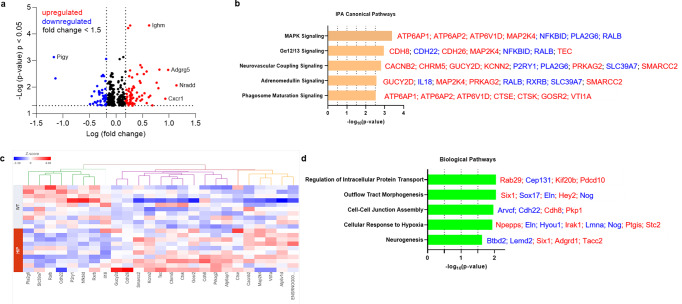
Fig. 7. The genes predicted to influence the impact of cerebrovascular amylin deposition on the central nervous system.
a The Log10 (p-value) versus Log10 (fold change) of the DE genes in HIP vs. WT rat brains. Each dot represents one gene, with upregulated in red color (HIP vs. WT, ≥ 1.5-fold change), downregulated in blue color (HIP vs. WT, ≥ 1.5-fold change), and < 1.5-fold change in black color. b Top 5 canonical pathways identified by Ingenuity Pathways Analysis of differentially expressed (DE) genes detected by RNA-seq analysis of hippocampal tissue from the HIP vs. WT rat groups (P < 0.05) (n = 10 males/group). c Hierarchical clustering of 25 DE genes identified the enrichment of top 5 canonical pathways in (b). d Top 5 Gene Ontology (GO) biological processes enriched in HIP rat group compared to WT rat group.