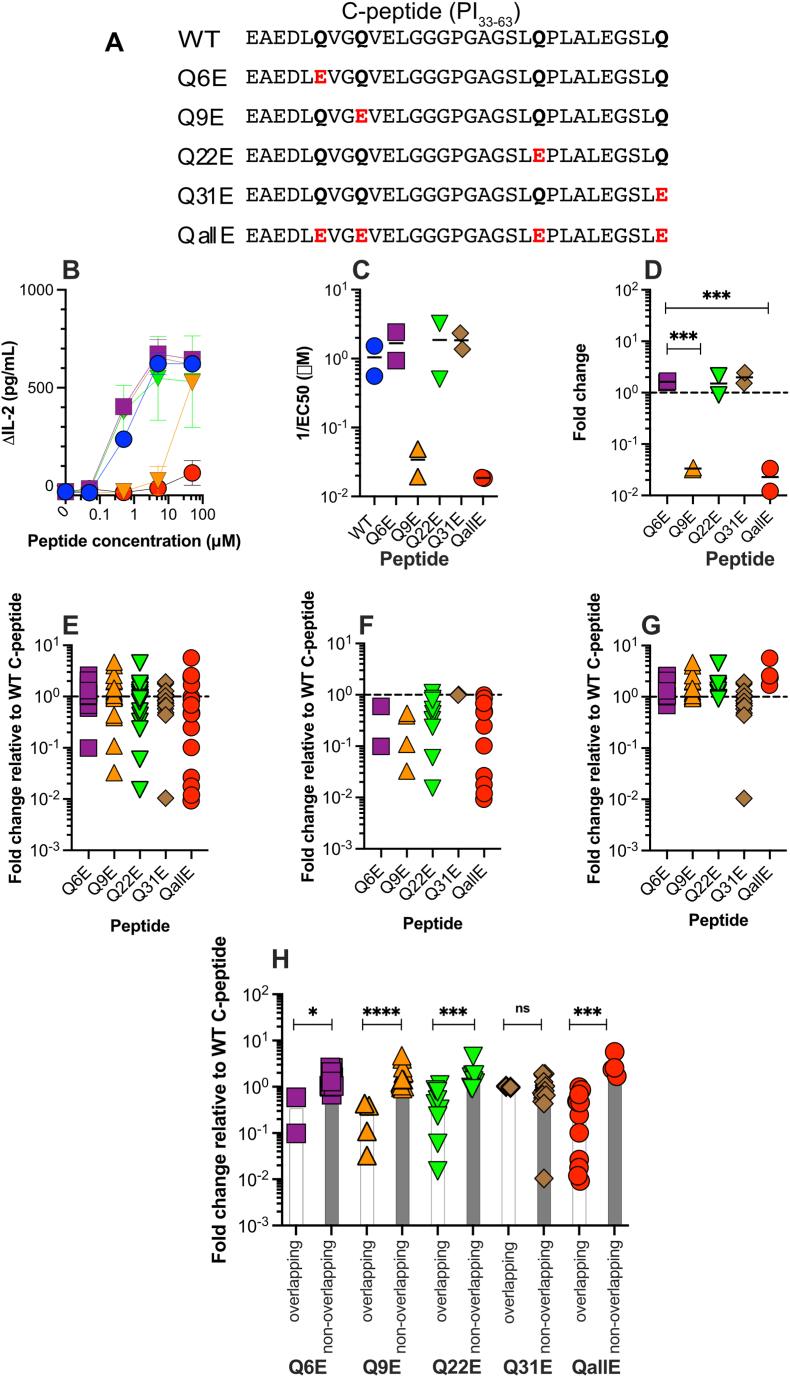
Fig. 2.
C-peptide specific TCR responses to deamidated C-peptide variants (A) Each peptide is referred to by the position, within C-peptide of the glutamine (Q), in bold, to glutamic acid (E), in red text, substitutions. The native sequence is referred to as wildtype (WT) C-peptide. The five variants are named according to the position within C-peptide of the substitution (i.e Q6E, Q9E etc), except for the peptide with all Qs substituted for E (QallE). (B) An example of a titration experiment using a Jurkat line (ACD4_38) expressing a C-peptide specific TCR. Each point represents the mean of triplicate measures and vertical bars, the standard deviation. (C) The EC50 for each peptide was calculated the and the EC50 from two independent experiments are shown. The bars represent the mean of the two experiments. (D) The fold change in EC50, relative to WT C-peptide for this TCR. The dotted line indicates a fold change of 1 relative to WT C-peptide. (E) A summary of the fold change, relative to WT C-peptide for all 18 clones tested. (F). Fold change for clones where the Q to E substitution falls within (F), or outside (G) the mapped epitope. (H) A comparison of the responses to a peptide when the modification falls within (overlapping) or outside (non-overlapping) the mapped epitope. Statistical significance was determined using the Mann-Whitney test (ns, not significant, 8, p < 0.05, **p < 0.01, ***p < 0.001).