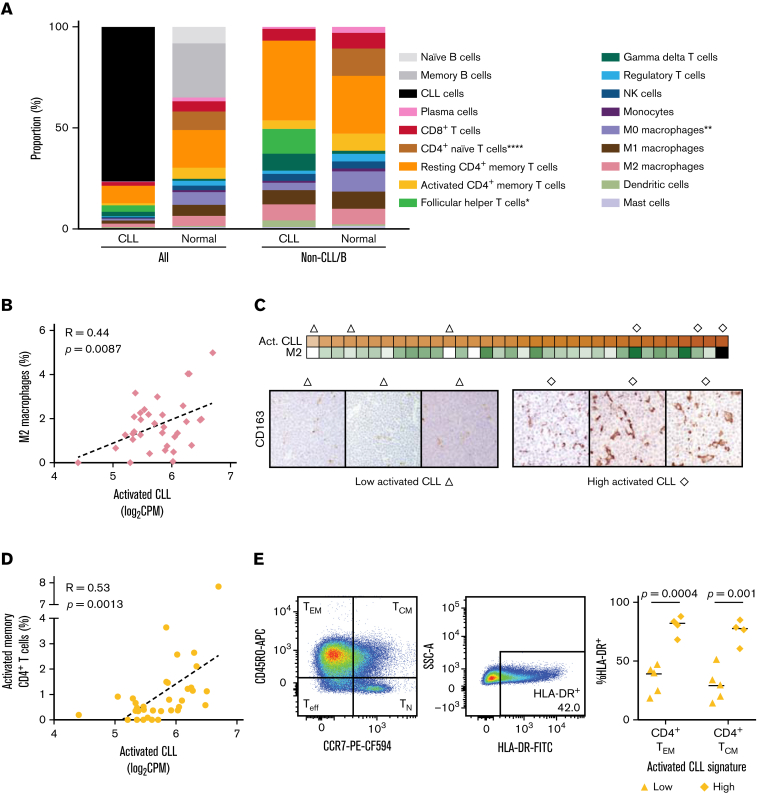
Figure 5.
Activated CLL cells interact with the immune microenvironment. (A) Mean abundance of the indicated cell types as estimated by CIBERSORT deconvolution of bulk RNA-seq data from CLL (n = 34) and normal LN samples (n = 4). There were significantly fewer follicular helper T cells (P < .05) and more CD4+ naïve T cells (P < .0001) and uncommitted macrophages in CLL LNs (P < .01) than normal LNs. (B) Correlation between the activated CLL signature and the abundance of M2 macrophages as estimated by CIBERSORT deconvolution of bulk RNA-seq data (n = 34). (C) Top: heatmap of activated CLL signature expression and CIBERSORT estimated abundance of M2 macrophages. Bottom: triangle and diamond symbols identify LN samples used for immunohistochemical staining of CD163+ M2 macrophages with low and high expression of the activated CLL signature. The average number of CD163+ cells per high-power field (40×) in each sample is provided immediately below. (D) Correlation between the activated CLL signature and the abundance of activated memory CD4+ T cells as estimated by CIBERSORT deconvolution of bulk RNA-seq data (n = 34). (E) Left: gating strategy of HLA-DR+ CD4+ effector memory (TEM CD3+CD19-CD14-CD4+CD8-CD45RO+CCR7-) and central memory (TCM CD3+CD19-CD14-CD4+CD8-CD45RO+CCR7+) cells in a representative flow cytometry dot plot. Right: proportion of CD4+ TEM and TCM cells that are HLA-DR+ in LN samples with low (n = 5) and high (n = 4) expression of the activated CLL signature.