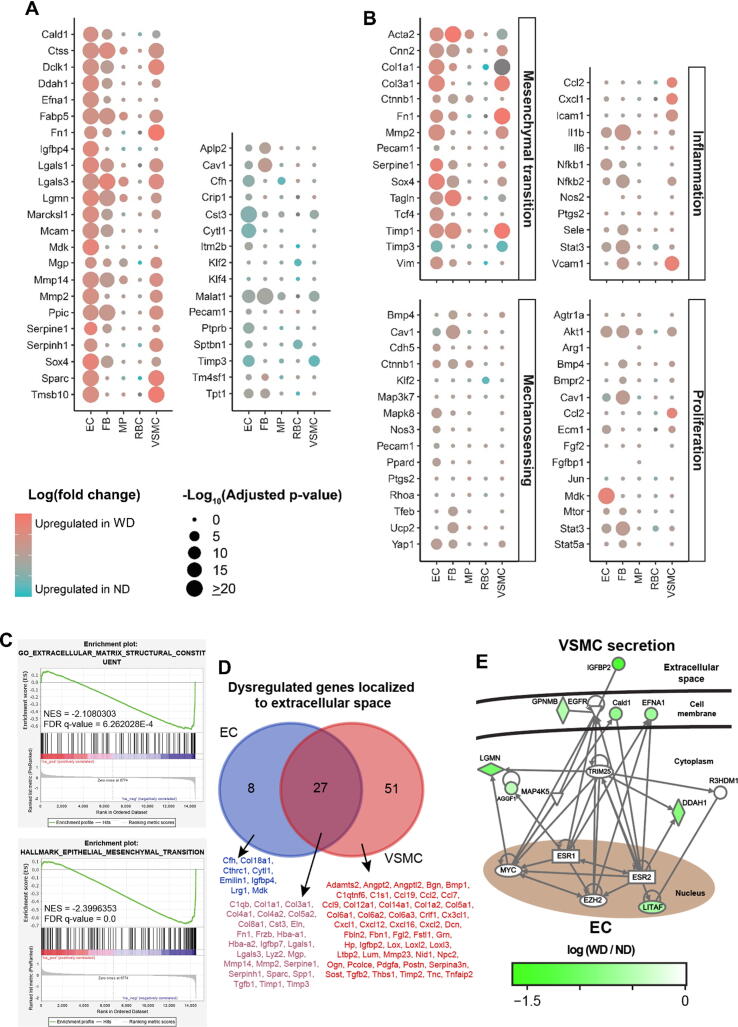
Fig. 2.
Transcriptional profiles of distinct cells in atherosclerotic aorta. (A) Dotplots overviewing selected up-regulated and down-regulated genes, and (B) highlighting expression of key genes in terms of mesenchymal transition, inflammation, mechanosening and proliferation. Dot colors, log (fold-change); size, adjusted p-value. (C) Gene set enrichment analysis (GSEA) on extracellular matrix structural constituent and mesenchymal transition of ECs between ND and WD groups. Upper plot: enrichment score distribution. Middle plot: black bars, genes of gene set; red and blue, downregulation and upregulation in WD group, respectively. Lower plot: ordered log (fold-change). Negative NES: enriched expression levels in WD group. (D) Venn-diagram on overlap of dysregulated genes localized to extracellular space of ECs and VSMCs, with the lists of common and unique genes. (E) Gene regulatory network analysis on the crosstalk between ECs and VSMCs during atherosclerosis. Scale bar: log (fold-change). EC, endothelial cell; FB, fibroblast; MP, macrophage; ND, normal diet; RBC, red blood cell; VSMC, vascular smooth muscle cell; WD, western diet. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)