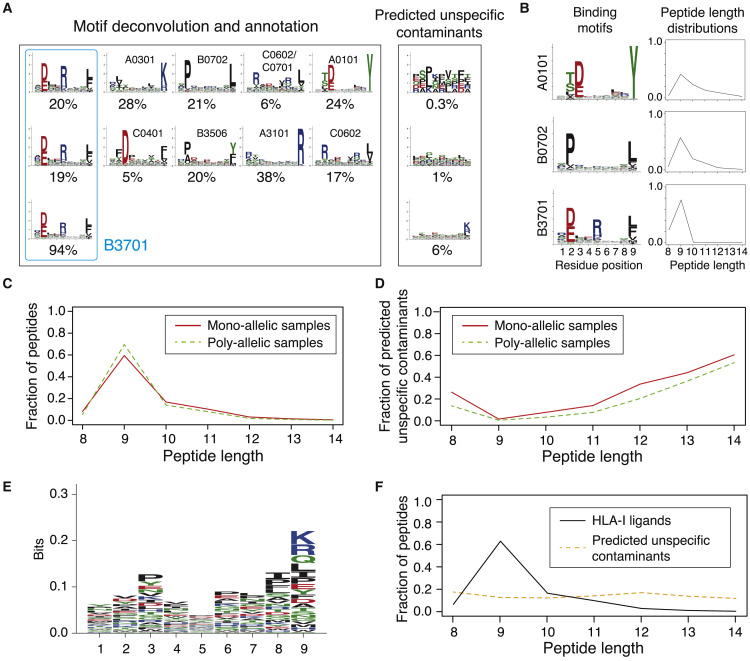
Figure 1.
Motif deconvolution across HLA-I peptidomics datasets reveal binding motifs and peptide length distributions of HLA-I molecules, as well as predicted contaminants in mono- and poly-allelic datasets.
(A) Example of motif deconvolution with MixMHCp in three HLA-I peptidomics samples. This includes determination of HLA-I motifs and predicted unspecific contaminants (i.e., peptides assigned to the flat motif of MixMHCp), as well as motif annotation by identifying shared motifs across samples sharing the same allele. The example shows the deconvolved motifs in two poly-allelic samples that share the HLA-B∗37:01 allele (“donor1” and “HCC1143” in Table S1), as well as the mono-allelic HLA-B∗37:01 sample.
(B) Examples of binding motifs and peptide length distributions obtained by motif deconvolution.
(C) Peptide length distributions in mono-allelic and poly-allelic HLA-I peptidomics data. Each curve represents the average peptide length distribution across alleles with both mono- and poly-allelic HLA-I peptidomics data.
(D) Fraction of predicted unspecific contaminants across different lengths (average over all samples).
(E) Motif of the predicted unspecific contaminants (9-mers) identified by MixMHCp across all samples.
(F) Comparison of the length distribution of peptides assigned to HLA-I alleles and predicted unspecific contaminants.