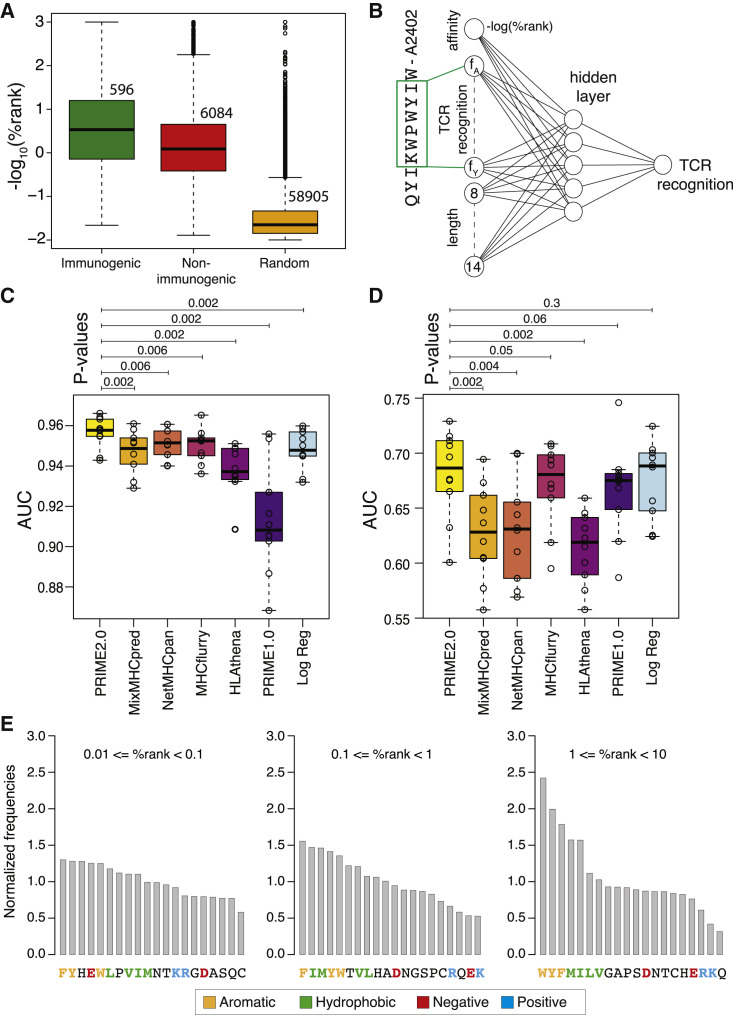
Figure 3.
Models of TCR recognition propensity improve predictions of neo-epitopes
(A) Predicted binding affinity to HLA-I (based on %rank of MixMHCpred2.2.) of experimentally validated immunogenic (green) and non-immunogenic (red) peptides, as well as random peptides (orange) used to train PRIME.
(B) Architecture of neural network of PRIME2.0. The first input node corresponds to the predicted binding to the HLA-I allele (−log(%rank) from MixMHCpred2.2). The next 20 nodes correspond to amino acid frequencies on residues with minimal impact on predicted affinity to the HLA-I allele (green box). These positions were determined as previously described.33 The last seven nodes correspond to the length of the peptide (i.e., 8–14, one-hot encoding).
(C) Benchmarking of PRIME2.0 based on 10-fold cross-validation. “Log Reg” indicates the model trained on the same data as PRIME2.0 but with a logistic regression instead of a neural network.
(D) Same cross-validation as in (C) after excluding randomly generated negatives in the test set.
(E) Normalized amino acid frequencies at positions with minimal impact on predicted affinity to HLA-I for immunogenic versus non-immunogenic peptides used to train PRIME2.0 within different ranges of predicted HLA-I binding (%rank of MixMHCpred).
Boxplots in (A), (C), and (D) represent the median and lower/upper quartiles. p values were computed with paired Wilcoxon test.