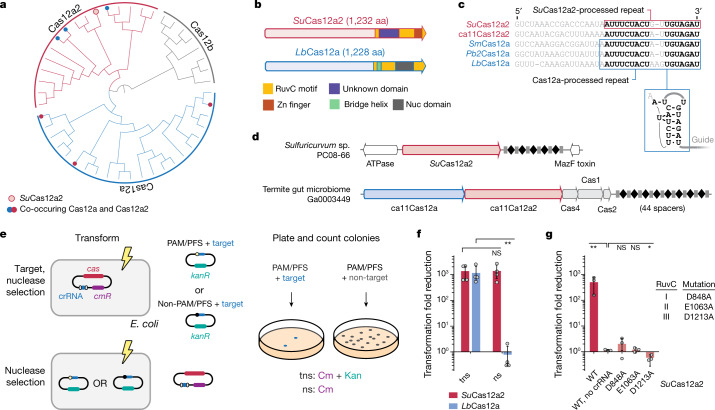
Fig. 1. Cas12a2 nucleases form a distinct clade within type V Cas12 nucleases.
a, Maximum-likelihood phylogeny of identified Cas12a2 nucleases with Cas12a and Cas12b nucleases. The detailed phylogeny is shown in Extended Data Fig. 1. Systems with co-occurring Cas12a2 and Cas12a are indicated by filled red and blue circles. SuCas12a2 is indicated by an unfilled red circle. b, The domain architecture of SuCas12a2 in comparison to LbCas12a. aa, amino acids. c, Aligned direct repeats associated with representative Cas12a2 and Cas12a nucleases. The bold nucleotides indicate conserved positions within the processed repeats for both nucleases. The predicted pseudoknot structure of the Cas12a repeat is shown below. The loop of the hairpin (grey) is variable. Pre-crRNA processing by SuCas12a2 is shown in Extended Data Fig. 3. d, Gene organization of CRISPR–Cas systems within representative genomic loci encoding Cas12a2. Examples of systems encoding Cas12a2 as the sole Cas nuclease and those also encoding Cas12a are shown. e, Diagram of the traditional (top; target nuclease selection (tns)) and modified (bottom; nuclease selection (ns)) plasmid interference assay. Cm, chloramphenicol; Kan, kanamycin. f, The reduction in plasmid transformation for SuCas12a2 and LbCa12a2 under target plasmid and nuclease plasmid selection. g, The reduction in plasmid transformation of SuCas12a2 RuvC mutants under target plasmid and nuclease plasmid selection. For f and g, data are mean ± s.d. of at least three independent experiments started from separate colonies. P values were calculated using one-tailed Welch’s t-tests; NS, P > 0.05; *P < 0.05, **P < 0.005.