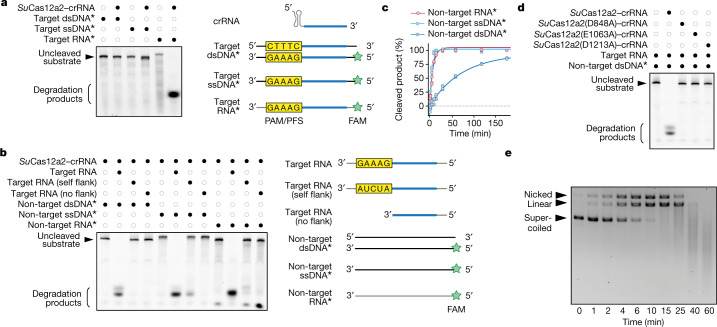
Fig. 2. RNA target recognition by SuCas12a2 triggers degradation of ssRNA, ssDNA and dsDNA in vitro.
a, Direct targeting of different FAM-labelled nucleic-acid substrates by a purified SuCas12a2–crRNA complex. b, Collateral cleavage of FAM-labelled non-target nucleic-acid substrates by the SuCas12a2–crRNA complex with different target RNA substrates after 1 h. Target RNA, a non-self flanking sequence at the 3′ end; self flank, a flanking sequence mutated to the reverse complement of the crRNA repeat tag; no flank, only the reverse complement of the crRNA guide. For a and b, diagrams of target and non-target nucleic acids are shown on the right. c, Time-course analysis of RNA-triggered collateral cleavage of labelled non-target RNA, ssDNA or dsDNA. Representative gel images are provided in Extended Data Fig. 4c. Note that dsDNA contains twice as much ssDNA substrate as the RNA and ssDNA, but the same concentration of labelled strands. d, The effect of mutating each of the three RuvC motifs on RNA-triggered collateral cleavage of dsDNA. e, Time-course analysis of RNA-triggered collateral cleavage of non-target plasmid DNA. Plasmid DNA was visualized using ethidium bromide. For a–d, the asterisks indicate a FAM-labelled substrate, and the diagrams on the right indicate the substrates. All of the results are representative of three independent experiments. Gel source data are provided in Supplementary Fig. 1.