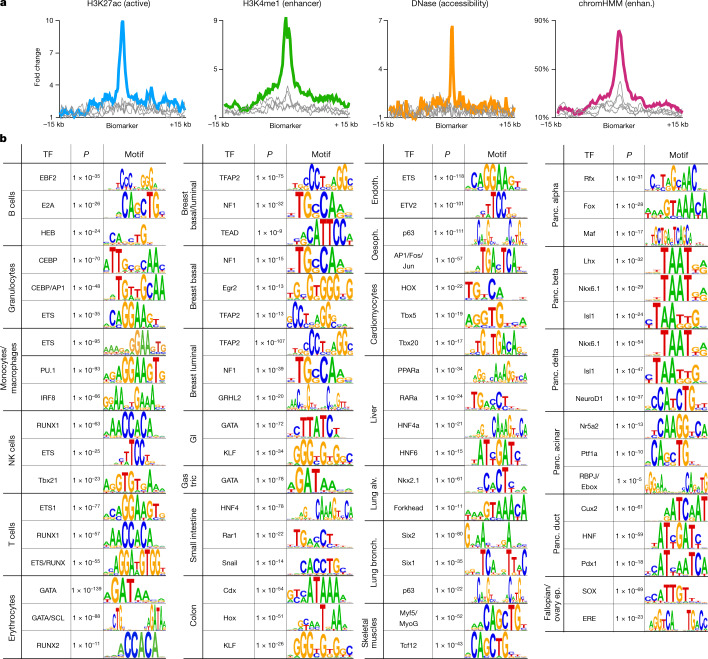
Fig. 4. Cell-type-specific markers as putative enhancers.
a, Average ChIP–seq signal for the active regulatory marker H3K27ac, enhancer marker H3K4me1, DNA accessibility and chromHMM enhancer annotations for the top 250 cell-type-specific unmethylated markers for monocytes/macrophages. Average signal for the top 250 markers of other blood cell types (granulocytes and B, T and NK cells) shown as grey lines, for comparison. b, Cell-type-specific markers are enriched for regulatory motifs. Shown are the top TF binding site motifs, enriched among the top 1,000 differentially unmethylated regions per cell type, using HOMER motif analysis. Motifs similar to previous (more significant) hits not included. Shown are HOMER binomial P values. Alv., alveolar; Bronch., bronchial; Endoth., endothelium; Ep., epithelium; Oesoph., oesophagus; Panc., pancreas.