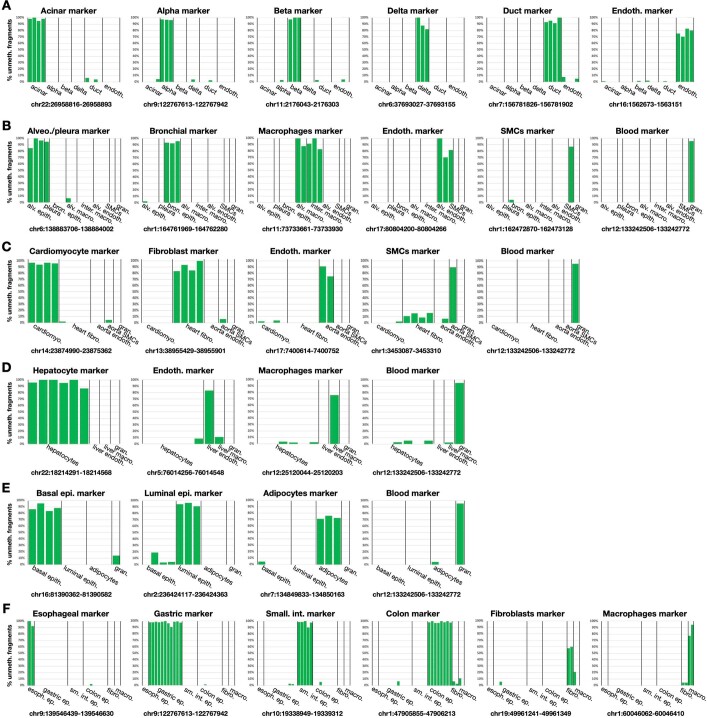
Extended Data Fig. 3. Purity estimation for pancreas, lung, heart, liver, breast, and GI using atlas markers.
The percent of unmethylated fragments (y-axis) among fragments of ≥4 CpGs from selected differentially methylation markers could serve as an (under-) estimate of the atlas purity. Here we show one such marker for each cell type, selected from the top 25 markers, and use fragment-level analysis to demonstrate the purity in the target cell type compared to other cell types from the same tissue or environment. (A) Pancreas. (B) Lung. (C) Heart. (D) Liver. (E) Breast. (F) GI tract. For most cell types, 90% of the molecules in the target cell types are unmethylated, compared with less than 5% of other types. This is an under-estimation, as some heterogeneity could occur in each cell type, reflecting stochastic noise, cellular states, age, or environmental changes.