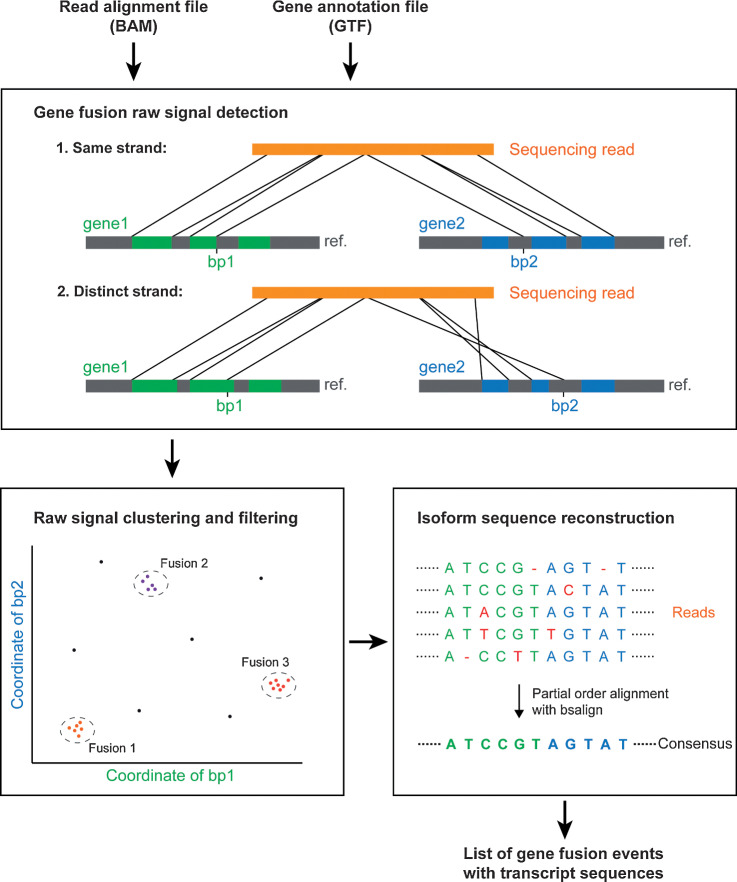
Figure 1.
Workflow of FusionSeeker. FusionSeeker scans the input file of the read alignments for split read alignments and records candidate fusions of gene fusions when two segments from one read are aligned to two distinct genes. It then clusters the candidate fusions into gene fusion calls and removes noise calls supported by only a few reads. For each fusion call, FusionSeeker generates a consensus transcript sequence by performing a POA with fusion-containing reads. The final output of FusionSeeker includes a list of confident gene fusion events and corresponding transcript sequences.