Figure 3 |. Errors corrected after polishing.
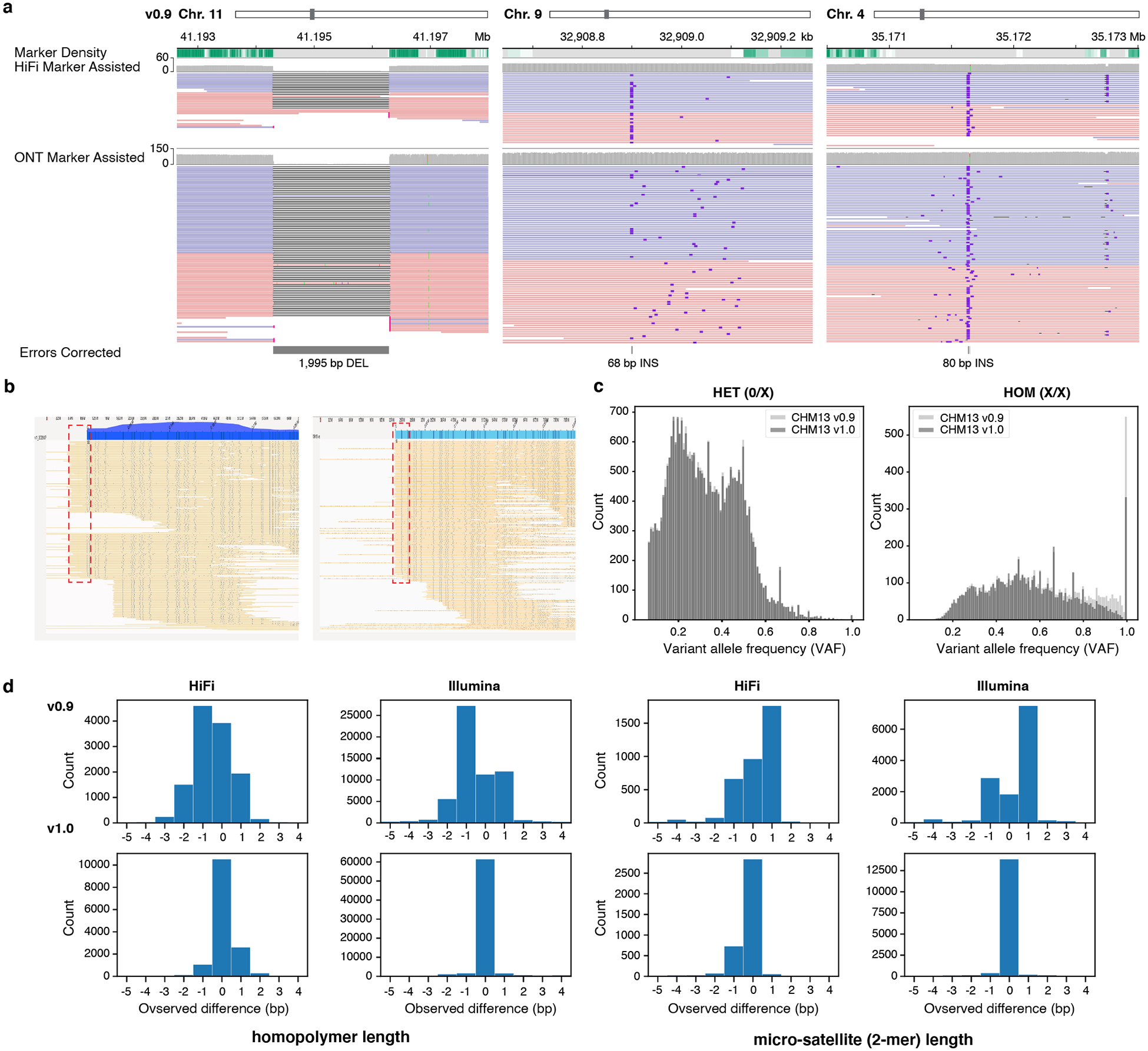
a, Three corrected SV-like errors. b, Bionano optical maps indicating the missing telomeric sequence on Chr. 18 p-arm (left) with a higher than average mapping coverage. This excessive coverage was removed after adding the missing telomeric sequence (right) and most of the Bionano molecules end at the end of the sequence. c, Variant allele frequency (VAF) of each variant called by DeepVariant hybrid (HiFi + Illumina) mode, before and after polishing. Most of the high frequency variants (errors) are removed after polishing, which were called ‘Homozygous’ variants. d, Total number of reads in each observed length difference (bp) between the assembly and the aligned reads at each edit position. Positive numbers indicate more bases are found in the reads, while negative numbers indicate fewer bases in the reads. Both the homopolymer and micro-satellite (2-mers in homopolymer compressed space) length difference became 0 after polishing.
