Extended Data Fig. 1 |. Sequencing biases observed in missing k-mers.
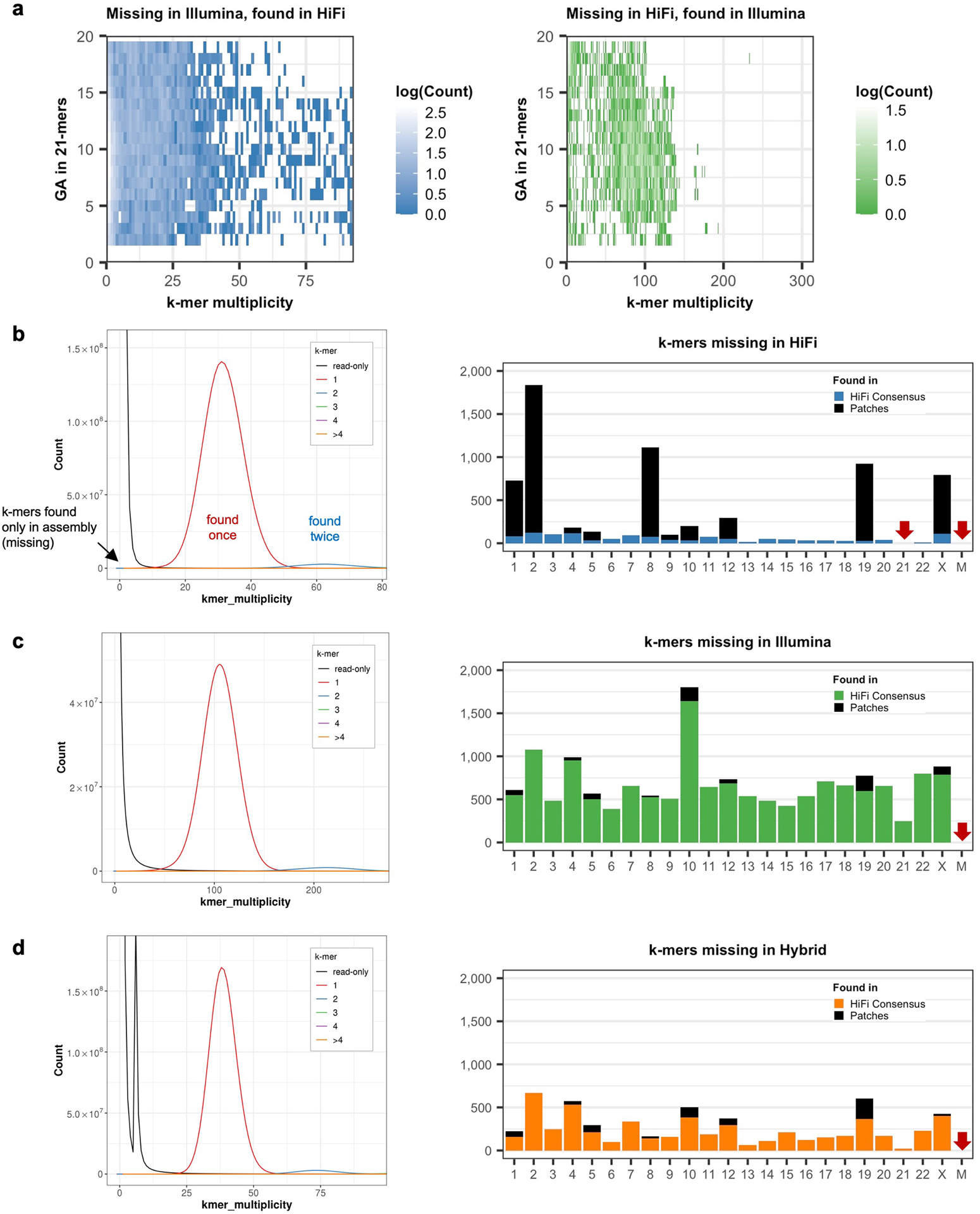
a, missing k-mers with its GA composition. b-d, v0.9 assembly and k-mer copy number spectrum from HiFi, Illumina, and hybrid k-mer sets (left) and per-chromosome missing (likely error) k-mer counts from the HiFi derived consensus or patches (right). Most missing k-mers in HiFi overlapped sequences from patched regions. No missing k-mer was found on chromosomes indicated with red arrows.
