Fig. 7. EpiDrivers function as Tumor Suppressors in Humans.
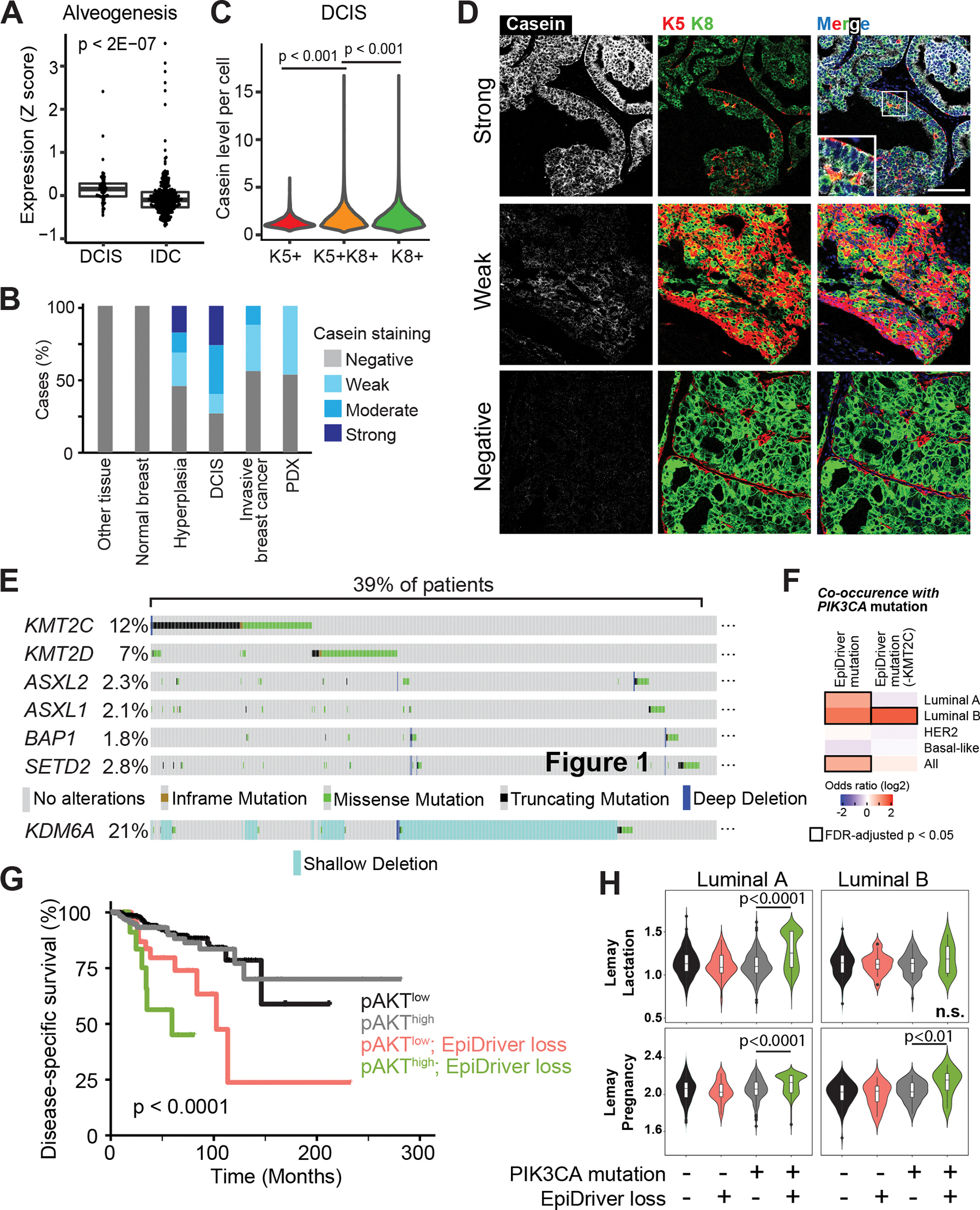
A, Average expression of the ‘Alveogenesis’ gene signature from 57 DCIS and 313 invasive tumors. B, Casein staining level by IHC in each tissue or tumor type. C, Casein staining intensity in individual cells in DCIS tumor cores separated by keratin staining. D, Representative imaging mass cytometry images of DCIS cores stained for casein and Krt5, Krt8 and nuclear stain. Scale bar = 100 μm. E, Prevalence of alterations in EpiDrivers in human breast tumors. Shallow deletion only displayed for KDM6A. F, Co-occurrence analysis of PIK3CA and EpiDriver mutations in the combined breast cancer dataset of TCGA and METABRIC. The results are shown for the complete set of identified EpiDrivers (left), or by excluding KMT2C (right), considering truncating and deleterious missense mutations. The heatmap shows the co-occurrence odds ratios (log2) across breast cancer subtypes, and all tumors considered, and significant (FDR-adjusted p < 0.05) associations are highlighted by black rectangles. g, Disease-specific survival (DSS) of breast cancer patients in the TCGA cohort stratified by phospho-Ser473 AKT (pAKT) and EpiDriver mutations. The long-rank p value is shown. h, Violin plots showing the expression of the Lemay Lactation and Pregnancy signatures in TCGA tumors with concurrent PIK3CA-EpiDriver mutations relative to other groups in luminal A and B breast cancer. The Mann-Whitney test p value is shown. The average value of the group with concurrent PIK3CA-EpiDriver mutations is depicted by a horizontal lilac line.
