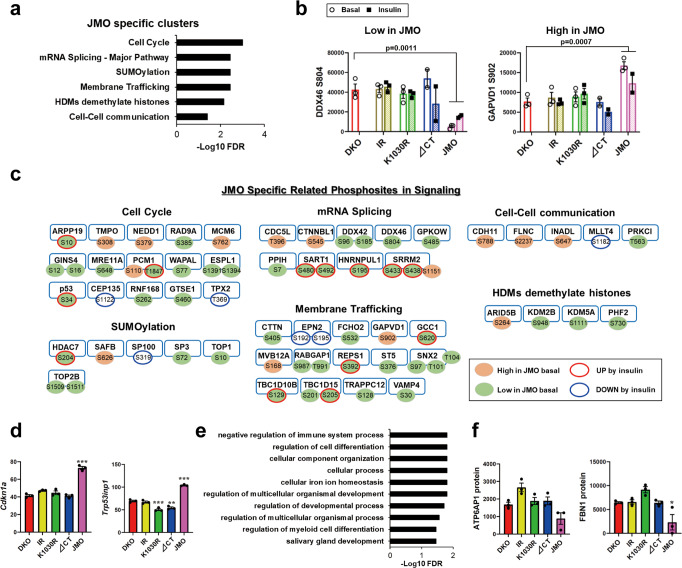
Fig. 7. JMO dependent signature in signaling and gene and protein expressions.
a REACTOME pathway enrichment analysis of phosphosites regulated by JMO-IR. The functional enrichment analysis was tested by the STRING database, where FDRs were calculated using the Benjamini–Hochberg procedure. Plots are −log10 transforms of enrichment FDR value. b Quantification of exemplary phosphosites in the enriched pathways (in (a)) in the “JMO specific” cluster. Data are means ± SEM of phosphosites intensity values (×104). P-values vs. DKO (combined both basal and insulin for comparisons), one-way ANOVA followed by Dunnett’s multiple comparisons test (n = 3–6). c Diagram of intracellular signaling regulated by JMO-IR as identified by phosphoproteomics. Each of the phosphosites was color-coded based on the effects of basal or insulin stimulation on phosphorylation. d Quantification of exemplary genes in the high expression in JMO cluster in the RNA-sequencing heatmap at the basal. Data are means ± SEM of genes intensity values (n = 3). **P < 0.01, ***P < 0.001 vs. DKO, one-way ANOVA. e, f The top 10 enriched GO term (Biological process) (e) and quantification of exemplary proteins (f) in the low protein expression in JMO cluster in the proteome heatmap at the basal. The functional enrichment analysis in (e) was tested by the STRING database, where FDRs were calculated using the Benjamini–Hochberg procedure. Data are means ± SEM of proteins intensity values (n = 3). *P < 0.05 vs. DKO, one-way ANOVA.