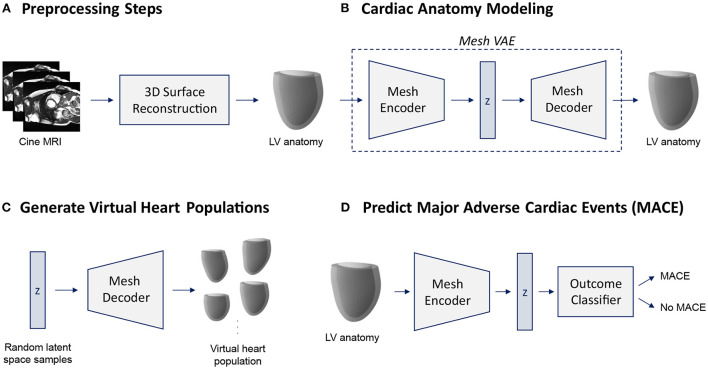
Figure 1.
Overview of the proposed cardiac shape analysis pipeline (A,B) and the two possible applications investigated in this work (C,D). First, we pass raw cine MRI acquisitions through a multi-step 3D surface reconstruction pipeline (Section 2.2) to reconstruct 3D surface meshes of the left ventricular (LV) anatomy as a preprocessing step (A). It consists of an image segmentation step using two cascaded U-Nets followed by a personalized template mesh fitting step based on variational warping techniques (Section 2.2). Next, as the core part of the pipeline, we propose a variational mesh autoencoder with an interpretable latent space z to efficiently capture population-wide variability in 3D cardiac shapes in a single geometric deep learning model (B) (Section 2.3). The decoder of the pre-trained mesh VAE can then be used to generate virtual population cohorts of 3D heart meshes (C) (Section 3.5). The pre-trained encoder converts input meshes into low-dimensional latent space representations which can serve as inputs to a MACE outcome classifier (D) (Section 3.6).