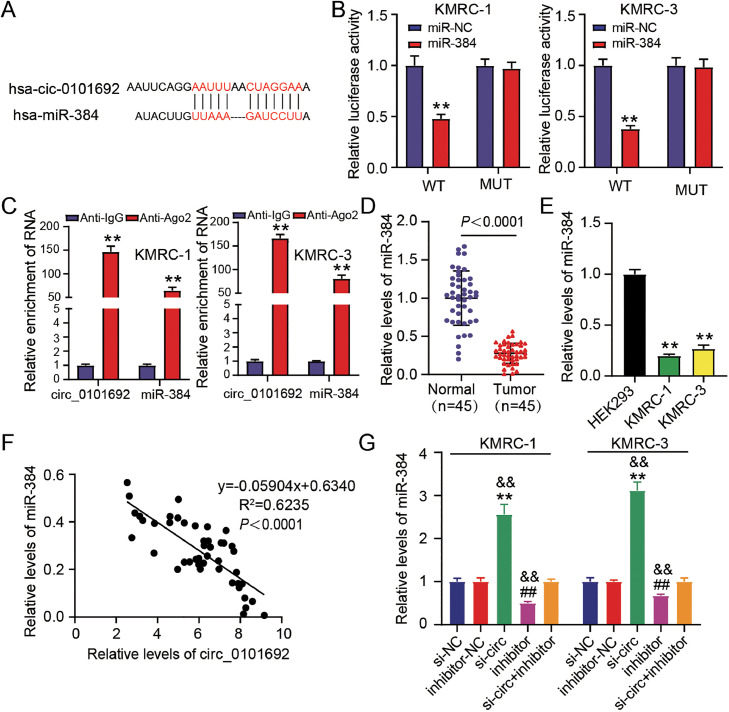
Fig. 3.
MiR-384 is a potential downstream target of circ_0101692. (A) The predicted binding site of circ_0101692 and miR-384 via circInteractome (https://circinteractome.nia.nih.gov/index.html). (B) The luciferase activity in co-transfected KMRC-1 and KMRC-3 cells with circ_0101692 WT (WT) or circ_0101692 MUT (MUT) and miR-384 mimic (miR-384) or miR-NC was ascertained by dual luciferase reporter assay. **P < 0.01 vs. miR-NC. (C) RIP assay was performed to validate the association among circ_0101692 and miR-384. **P < 0.01 vs. Anti-IgG. (D) The expression of miR-384 in ccRCC tissues and normal tissues was detected by qRT-PCR. P < 0.0001 vs. Normal. (E) The expression of miR-384 in ccRCC cell lines (KMRC-1 and KMRC-3) and HEK293 cells was ascertained with the help of qRT-PCR. **P < 0.01 vs. HEK293. (F) Correlation between the expression of circ_0101692 and miR-384 in ccRCC tissues was assessed by Pearson correlation analysis. (G) The expression of miR-384 in KMRC-1 and KMRC-3 cells after transfection si-circ, si-NC, miR-384 inhibitor (inhibitor), inhibitor-NC, or si-circ + inhibitor was detected by qRT-PCR. **P < 0.01 vs. the si-NC group; ##P < 0.01 vs. inhibitor-NC; &&P < 0.01 vs. si-circ + inhibitor.