Abstract
The variable domain of new antigen receptor (VNAR) of shark single domain antibodies is evolutionarily distant from the variable regions (VH) of mammalian immunoglobulins, yet they still have complementarity-determining regions (CDRs) that are involved in antigen recognition, therefore making it possible to humanize them by grafting these CDRs to the framework of human VH homologs. Here we show the VNAR CDR based on an analysis of currently available VNAR-antigen structure complexes in the global Protein Data Bank archive of 3D structure data and describe the detailed protocol to humanize VNAR by CDR grafting, using B6 (an anti-pseudomonas exotoxin VNAR), the most common type (Type II) of shark VNARs, as an example. Ongoing efforts will further optimize the protocol for moving shark VNARs to the clinic for treating cancer and other human diseases.
Basic Protocol 1: humanize Shark VNAR sequence by CDR grafting
Support protocol 1: VNAR structure prediction and comparison
Support protocol 2: measure binding kinetics of humanized VNAR using bio-layer interferometry (BLI)
Keywords: Shark VNAR single domain antibody, humanization, CDR grafting
INTRODUCTION:
In comparison to conventional immunoglobulin antibodies, cartilaginous fish such as sharks, rays, and skates possess a unique form of antibody that consists of a homodimer of two heavy chains only. This unique antibody form is termed immunoglobulin new antigen receptor or IgNAR in its short form (Flajnik, 2018; Greenberg et al., 1995; Hamers-Casterman et al., 1993). Each heavy chain of this unique immunoglobulin is comprised of five constant domains following a single variable domain (V domain of IgNAR, named VNAR), which functions as one of the most ancient antigen recognition receptors (English et al., 2020a; Stanfield et al., 2004; Streltsov et al., 2004; Zielonka et al., 2015). The main purpose of humanizing shark VNARs is to develop them into therapeutic molecules. Antigen-specific shark VNARs are usually isolated from phage display libraries, either from immunized or antigen-naïve sharks. We generated a large antigen-naïve VNAR library from six nurse sharks (Ginglymostoma cirratum) with a diversity of 1011 (Feng et al., 2019). The B6 VNAR used in this protocol was isolated from our shark VNAR library (Feng et al., 2019). Because shark VNARs have unique structures and binding modes that are different from those of mammalian antibodies (Dooley et al., 2006; Stanfield et al., 2004), VNARs may have unique therapeutic potential (English et al., 2020b). CDR grafting is the most straightforward and widely used humanization approach (Carter et al., 1992; Jones et al., 1986; Queen et al., 1989): CDRs of a non-human antibody of interest are grafted onto an appropriate human germline framework and retain the binding and functional property of the original antibody. Early work on shark VNAR humanization based on Type I and Type IV was carried out by Kovalenko et al. (Kovalenko et al., 2013) and further explored by Steven et al. (Type IV VNAR) (Steven et al., 2017). To do CDR grafting, we need to determine where are the CDRs and what human template is suitable to accommodate the shark CDRs. CDRs were defined by sequence variability (KABAT) (Kabat et al., 1992; Wu et al., 1970) or structure features (Chothia et al., 1987) in early days, and by the identification of antigen-contacting residues when more antibody-antigen complex structures were available (Kunik et al., 2012; Padlan et al., 1995). We previously defined the antigen contacting residues in the rabbit monoclonal antibodies based on the analysis of the structures available in the global Protein Data Bank (PDB) and used them to guide rabbit antibody humanization (Zhang et al., 2017). In this article, we show shark antigen-contacting residues in a nonredundant shark VNAR structure database of nine antibody-antigen pairs and validate whether Kovalenko’s method can be adapted to humanize Type II VNARs. The next-generation sequencing analysis of a VNAR repertoire based on our large phage-displayed library from six naïve nurse sharks shows that Type II is the most common shark VNAR (> 70%) among all the types (Type I – IV) (English et al., 2020b; Feng et al., 2019). Here, we provide this humanization protocol as the basic protocol to humanize a Type II shark VNAR with an unknown structure via CDR grafting. The output of the basic protocol is the humanized VNAR sequence as a starting point for further sequence optimization. We also provide a supporting protocol to create a structural model of the shark and humanized VNARs and show how to superimpose them to check the humanization. Researchers can then produce the VNAR protein from the humanized VNAR sequence in E. coli using our published protocol (Z. Duan et al., 2022), and measure binding kinetics of the protein using Octet using the supporting protocol 2 described here.
Basic protocol: humanize shark VNAR sequence by CDR grafting
This part describes how to convert shark VNAR sequence into humanized VNAR sequence. We use our anti-pseudomonas exotoxin VNAR clone B6, a Type II VNAR isolated from our phage library of nurse shark VNARs (Feng et al., 2019). We will first align the shark sequence with human template DPK9-JK1 (Kovalenko et al., 2013), and then graft the shark sequences to the human template. The resulting sequence can be checked by structure modeling and model comparisons, and can be produced as VNAR proteins.
Unlike conventional heavy chain variable domains, the distinct immunoglobulin-like structure of VNAR domains possesses only two major complementarity determining regions (CDR1 and CDR3). The CDR2 region is replaced by a short β-strand formed hyper variable domain 2 (HV2) (Stanfield et al., 2004; Streltsov et al., 2004). An additional hypervariable region is called HV4 (Dooley et al., 2006). The HV2 occupies a similar space for CDR2. HV4 occupies a similar space to LV4 (the fourth loop besides CDR1, 2, 3 that may contact antigen (Zhang et al., 2017)) in conventional antibodies (Kovalenko et al., 2013). Mutational analysis of IgNAR 5A7 shows that IgNAR is greatly tolerant to amino acid substitutions in several areas including framework 1 (FW1), CDR1, parts of framework 2 (FW2), and both HV2 and HV4 regions, and a few residues outside CDR1 and CDR3 may also play a role in binding to the antigen (Fennell et al., 2010). There are four types of shark VNARs (Type I – IV) according to the number and location of disulfide bonds (English et al., 2020b) (Figure 1).
Figure 1.
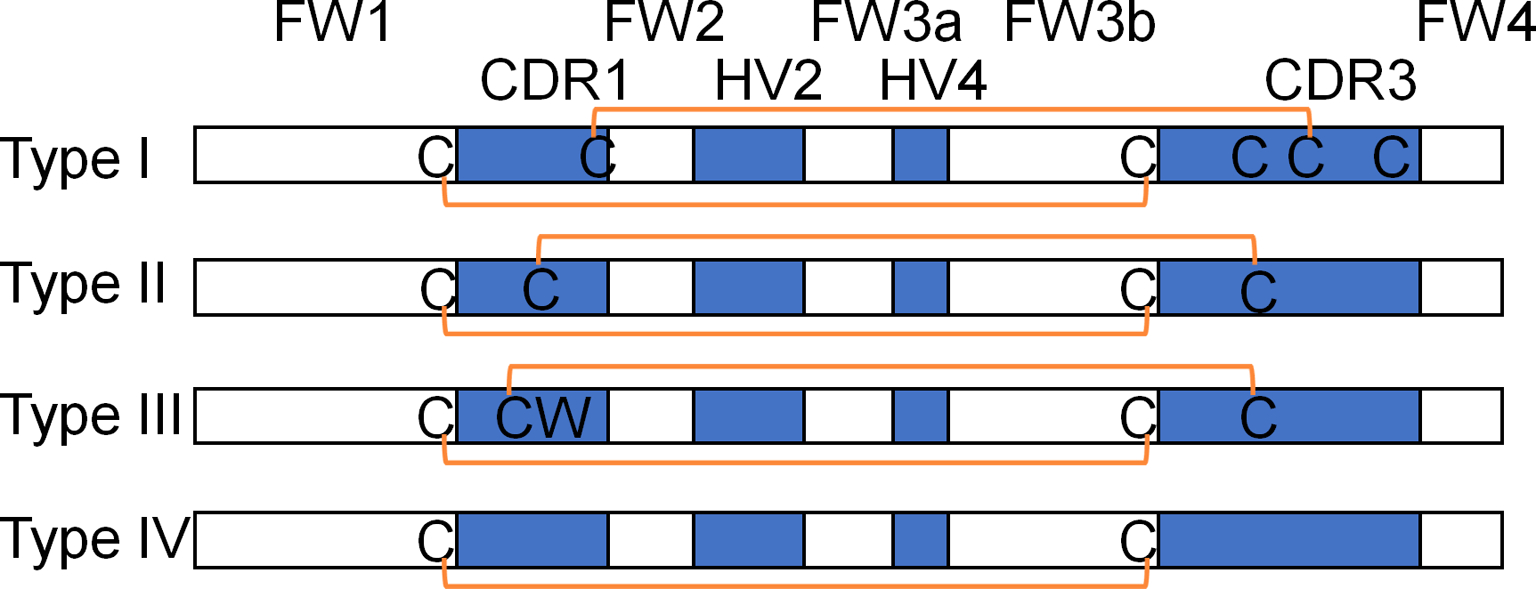
A) Four types of shark VNAR based on the location and number of disulfide bonds. The blue boxes show the location of CDR1, HV2, HV4 and CDR3. The orange lines show the disulfide bond.
To help the readers understand the principle of the shark VNAR humanization, we need to first show the antigen-contacting residues on shark VNARs. To identify shark VNAR CDR, we first generated a non-redundant shark VNAR-antigen complex structure database from published structures in the PDB. As of September 5, 2022, there are nine non-redundant VNAR-antigen complex structures in the PDB database, two type-I, six type-II, and one type-IV VNAR. They are from four kinds of shark species: Ginglymostoma cirrature, Orectolobus maculatus, Squalus acanthias, Chiloscyllium plagiosum. We aligned the structures using Mustang multiple protein structural alignment algorithm (http://lcb.infotech.monash.edu.au/mustang/) and identified the antigen-contacting residues using CCP4 with the CONTACT/ACT program(https://www.ccp4.ac.uk/)(Winn et al., 2011), as we previously did (Zhang et al., 2017). We consider two residues at the closest atom pairs no more than 6 Å apart as contacting residues, as this is the same criteria originally used by Kunik et al.(Kunik et al., 2012) to define the human and mouse antibody antigen-binding residues. We then process the data using excel and highlight the antigen contacting residues on the structure alignment. The multiple structure alignment and antigen-contacting residues are shown in Figure 2 and S1. This is the guideline that we use to humanize shark VNAR.
Figure 2.

Multiple structure alignment of VNARs from the PDB database. A) Sequences. B) Structures. These VNARs were extracted from the VNAR-antigen complex structure using PyMOL, and aligned using Mustang. The antigen-contacting residues are identified based on the close proximity (≤ 6 Å) of at least one atom in the antibody residue to the closest antigen atom. KABAT numbering at landmark positions are labeled on the sequence according to the human DPK9-JK1 alignment shown in Figure 3. Antigen-contacting residues are concentrated in CDR1, FW2, HV2-FW3a-HV4, and CDR3 regions. Therefore, we may be able to graft the VNAR CDR from various VNAR types to a common human framework.
Materials:
A general “wet laboratory” computer (either PC or Macintosh) with internet access for sequencing analysis.
Reagents
Competent cells E. coli strain HB2151 (LifeScienceMarket, cat. no. S0122) or Expi293F (ThermoFisher Scientific, cat. no. A14527)
Phagemids: pComb3X-S3B3; pComb3X-CG2G2; pComb3X (a gift from Dennis Burton at the Scripps Research Institute, La Jolla, CA; available from Addgene, cat. no. 63888); pcDNA4/TO (ThermoFisher Scientific, cat. no. V102020
2XYT medium (Research Products International, cat. no. L24045)
SOC medium (ThermoFisher Scientific, cat. no. 15544034)
50% (v/v) Glycerol (Sigma-Aldrich, cat. no. G5516)
100 mg/ml Ampicillin (Sigma-Aldrich, cat. no. A1593)
2XYT agar plates with 100 μg/ml ampicillin and 2% (w/v) glucose
20% (w/v) glucose solution (Sigma-Aldrich, cat. no. D9434)
Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich, cat. no. 16758)
Polymyxin B (Sigma-Aldrich, cat. no. P0972)
Protease inhibitor cocktail 100X (ThermoFisher Scientific, cat. no. 78429)
Buffer A: PBS + 0.5 M NaCl
Buffer B: Buffer A + 200 mM imidazole (Sigma-Aldrich, cat. no. 68268)
Phosphate-buffered saline (PBS) 10x (ThermoFisher Scientific, cat. no. 70011044)
PBST: 1x PBS containing 0.1% (v/v) Tween 20 (Biorad, cat. no. 1610781)
Deionized water
HisTrap HP His tag protein purification column 1 × 1 mL (Cytiva, cat. no. 29051021)
SDS-PAGE gel: Novax 4%−20% Tris-Glycine gel (Invitrogen, cat. no. XP04205BOX)
Precision Plus Protein™ All Blue Prestained Protein Standards (Biorad, cat. no. 1610373)
4x Laemmli sample buffer (Biorad, cat. no. 1610747)
TMB Chromogen Solution (ThermoFisher Scientific, cat. no. 002023)
0.45-μm syringe filter (Millipore, cat. no. SLHVN33RS)
MaxiSorp 96-well plates (Sigma-Aldrich, cat. no. M9410)
Ice
Equipment
AKTAexplorer (GE Healthcare Life Sciences)
Nanodrop 1000 spectrophotometer (ThermoFisher Scientific)
Centrifuge with 96-well microtiter plate adapter (Sorvall)
Spectrometer (Amersham GE Healthcare)
Incubator shaker (New Brunswick Scientific) with clamps
Magnetic stirrer (ThermoFisher Scientific)
Slide-A-Lyzer dialysis cassette 10K (ThermoFisher Scientific, cat. no. 66380)
250-ml Erlenmeyer flask (Sigma-Aldrich, cat. no. CLS431144)
2-liter Erlenmeyer flask (Sigma-Aldrich, cat. no. CLS431255)
L-shaped cell spreader (CellTreat, cat. no. 229617)
Protocol steps with step annotations:
To humanize shark VNAR, follow this procedure:
Align the sequences using Clustal omega (https://www.ebi.ac.uk/Tools/msa/clustalo/), Input the user-provided protein sequences of shark VNAR (for example, B6 sequence: ARVDQTPQTITKETGESLMFTCVLRDSNCGLSRTFWYRTKTGSTNEERISKGGRYVETV NSGSKSFSLRINDLTVEDSGTYRCRGKSWYYECPYDVYGGGTVVTVN) and human framework DPK9-JK1 (DIQMTQSPSSLSASVGDRVTITCRASQSISSYLNWYQQKPGKAPKLLIYAASSLQSGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQSYSAPRTFGQGTKVEIK) in FASTA format (this is input format), to align the two sequences. For demonstration purposes, we also put in previously humanized Type I VNAR 5A7 (PDB: 1SQ2 chain N) and Type IV VNAR E06 (PDB: 4HGK chain C) and their humanized versions (Figure 3A, S2).
- Highlight the grafted sequence as shown in Figure 3A in the aligned sequence. In the VNAR sequence, do the following:
- Identify the canonical C (Cys-23, KABAT position 23), which marks the end of FW1, and the next amino acid residue is the start of CDR1.
- Identify the canonical W (Trp-35), which marks the start of FW2, and the previous amino acid residue is the end of CDR1.
- Identify the canonical 41G (Gly-41), and the HV2 will start from position 43.
- Identify the canonical 70S (Ser-70), which marks the end of HV4.
- Identify the canonical 88C (Cys-88), which marks the end of FW3, and the CDR3 will start with the next amino acid residue.
-
Identify 101G (Gly-101), which marks the start of framework 4.
Copy and paste the CDR1, KABAT position 37, 38, 39, HV2-FW3a-HV4, and CDR3 sequences from VNAR into the human DPK9-JK1 framework as shown in Figure 3A.
Synthesize the humanized sequences and clone it into an expression vector. Many vendors offer gene synthesis and cloning service. Make sure to include signal peptide sequence in the expression vector or preceding the synthesized sequence (Zhijian Duan et al., 2022). Express the protein and compare the binding ability of humanized VNAR to the parental VNAR by following our recently published protocol (Zhijian Duan et al., 2022). Briefly, for E. coli expression, transform VNAR in the pComb3x vector into competent HB2151 cells, culture to an OD600 of 0.8, then induce expression by IPTG addition in the absence of glucose, and the VNAR protein will be secreted into the bacterial periplasmic space and can be released by polymyxin B treatment. Finally, purify the secreted VNAR by using a HisTrap column. In the case of B6, the yield of hB6 was 8.6 mg and the yield of B6 was 3.4 mg from 500 ml bacteria culture. The hB6 showed better soluble expression than B6. For mammalian cell expression, the VNAR can be cloned into a mammalian expression vector such as pcDNA4 with a signal peptide, and transfected into Expi293F cells; it will then be secreted into the culture medium, and can be purified using a HisTrap column. In Figure 4A, we show the SDS-PAGE of the purified VNAR. We prefer the bacteria expression system because it is an economic way of VNAR production.
Figure 3.
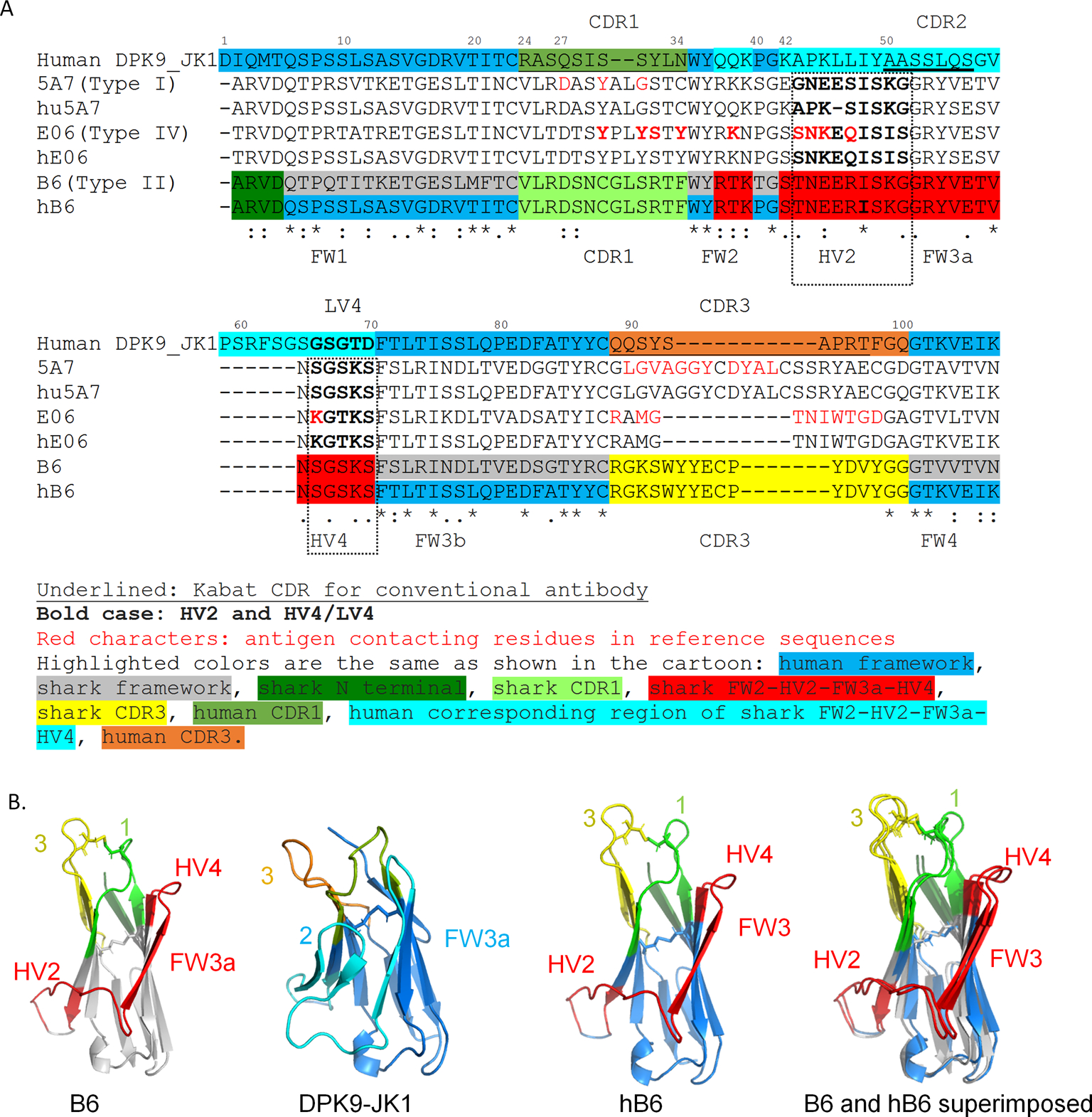
Shark type II VNAR B6 humanization by CDR grafting. A) Sequence alignment of B6, CDR grafted humanized B6 (hB6), human germline sequence DPK9_JK1, shark VNAR 5A7, humanized 5A7, VNAR E06, and humanized E06. Amino acid sequence of human germline antibody DPK9_JK1 was used as a template. The N-terminal CDR1, part of FW2 (KABAT position 37, 38, 39), HV2-FW3a-HV4, and CDR3 of VNAR were grafted from shark VNAR. Shark VNARs 5A7 and E06 are previously reported humanization references. KABAT numbering according to human sequence was used. Because the VNAR does not have a HV2, there is a gap between positions 58 and 65 in VNAR. B) The structure model of B6, humanization template DPK9-JK1, humanized B6 (hB6), and the superimposition of B6 and hB6. Structure models were built by “ColabFold: AlphaFold2 using MMseq2” (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb) (Jumper et al., 2021). Structure models were drawn with PyMOL, the superposition was made with the Dali server. The sidechains of all cysteines are shown as sticks. The disulfide bond between B6 CDR1 and CDR3 is formed in the protein models and is kept in the hB6.
Figure 4.

Biophysical and functional characterization of B6 and hB6. A) After elution off HisTrap column, B6 and hB6 (both are His-tagged) proteins in the collected fractions A15, B15, B14 were analyzed by SDS-PAGE. B) The binding kinetics of B6 and hB6 measured by octet. B6 and hB6 (both are His-tagged) were immobilized on Ni-NTA biosensors to bind free PE38. The association and dissociation curve of PE38 is shown. The Y-axis measures wavelength change in biolayer interferometry peaks (in nm), which is a function of changes in average optical thickness and correlates with ligand binding.
SUPPORT PROTOCOL 1: VNAR structure prediction and comparison
This protocol describes how to use Alphafold2 to predict VNAR structures from protein sequences and subsequently align and generate superimposed views of the structure models of shark VNAR and humanized VNAR. One will be able to generate a figure like Figure 3B.
Additional Materials:
Protein structure visualization software, such as PyMOL (https://pymol.org/2/).
Protocol steps with step annotations:
Open a web browser such as Chrome, go to the https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb, sign in to a google account (one can sign in by providing an email address). Input the protein sequence of shark VNAR or humanized VNAR into the “query_sequence”. One can check “use_amber” to run additional relaxation. The default value of other fields can be kept.
Click “Runtime” to open the dropdown menu, select “run all”, then wait. While waiting, please make sure the computer does not fall asleep to avoid unexpected termination of the computing. It could take more than half an hour to finish the computing.
After the run, Chrome will automatically download the result. Open it and find the .pdb file with “rank_1” in its name. This is the best model for this single domain sequence query, ranked according to the predicted local distance difference test (pLDDT)(Tunyasuvunakool et al., 2021). The pLDDT is set on a scale from 0 to 100, where 100 indicates the most accurate prediction. It estimates how well the prediction would agree with an experimental structure based on the local distance difference test Cα (lDDT-Cα). Regions with pLDDT > 90 are expected to be modeled to high accuracy, regions with pLDDT between 70 and 90 are modeled well (https://alphafold.ebi.ac.uk/faq). In this example, B6 pLDDT score is 96.3, hB6 pLDDT score is 96.7.
Align the structure models of the shark VNAR and humanized VNAR, using DALI server (http://ekhidna2.biocenter.helsinki.fi/dali/). Go to the link, select the “Pairwise” tab, then upload the best structure models (.pdb files from step 3) of shark VNAR and humanized VNAR, click “Submit”.
When the result is ready, click “Interactive (html)”, in the “Summary” section one can view the Z scores, right click the link “PDB”, and select “Save link as” to download the file. Open the download folder, and change the file type of this downloaded file into a .pdb file by typing in the file name to change the extension.
Download protein structure visualization software, such as PyMOL (https://pymol.org/2/). Open the .pdb file using PyMOL to view the aligned structures. Click Display > sequence in PyMOL to view the sequence of the aligned structure, and determine whether the sequence belongs to shark VNAR or humanize VNAR. If the aligned sequence is shark VNAR, drag the humanized VNAR.pdb file into PyMOL and vice versa. PyMOL will then show the superimposed structures.
Check the location of the disulfide bonds by clicking on the sequences in PyMOL (https://pymol.org/2/) and show the amino acid cysteines (C) as sticks. Check the location of CDR1, HV2, HV4, and CDR3 by selecting these sequences and show them in a different color. A good humanization design should not greatly change the fold. If one wants to compare different versions of humanization, the superimposed models of different versions of humanized sequence with the shark VNAR can intuitively help one to choose the better version. One can also check the Z score in Dali alignment, higher Z-score means better alignment or more similar structures (Holm, 2020). Lower RMSD scores are more desirable than higher RMSD scores. In the Dali alignment of B6 and humanized B6, we got a Z-score of 20.8, and RMSD of 0.9 (Å).
SUPPORT PROTOCOL 2: measure binding kinetics of humanized VNAR using bio-layer interferometry (BLI).
This support protocol describes how to measure binding kinetics of VNAR using Octet equipment. [Octet can real-time measure the association and dissociation of antibody/antigen, and can calculate the KD values of antibodies as an indication of affinity (Sultana et al., 2015).] For specific instructions on how to operate Octet, please refer to the Octet application notes. In this protocol, we will load the VNAR on the Octet probe, and the monovalent antigen will be in the solution phase.
Additional Materials:
Purified shark VNAR and humanized VNAR protein with a His-tag (produced in lab. See Basic Protocol)
Antigen (no His-tag) specific for VNAR. In the case of our VNAR B6, the recombinant Pseudomonas exotoxin (LH7-PE38) is produced in our lab (N. Li et al., 2017; Pastan et al., 2010)
Assay buffer (see recipe in Reagents and Solutions)
Ni-NTA probe (Fortebio, cat. no. 18–5101)
Flat bottom black 96-well plate (Greiner Bio-One, cat. no. 655209)
Equipment: octet (previous Fortebio, now Sartorius, cat. no. OCTET RED96E)
Prewarm the octet instrument to 30 °C. Presoak the Ni-NTA probe in assay buffer in the instrument for 10–30 minutes. If using the recommended 96-well plate, octet need 200 μl/well solutions.
Prepare the VNAR in assay buffer at the predetermined loading concentration (see commentary for how to determine loading concentration). Prepare antigen in assay buffer at various concentrations (eg. 1nM ~ 100 nM) by serial dilution for antigen association phase. For each antigen concentration, set up a control with no antibody loaded on the probe. In addition, set up a control with no antigen (0 nM antigen).
Run the following kinetic program: 10 minutes presoak, 180 seconds baseline establishment, 300 seconds VNAR loading, 60 seconds baseline establishment, 600 seconds antigen association, 30 minutes or more antigen dissociation.
At the data analysis stage, for each antigen concentration, subtract its control with no antibody, then subtract the control with 0 nM antigen,. Calculate the KD using global fit of the subtracted curve. If the octet analysis software cannot subtract controls accordingly, align the curves in octet analysis software and then export the data; subtract the controls in excel and then analyze the subtracted data in prism. In Figure 4B, we show the result of humanized B6 validation.
REAGENTS AND SOLUTIONS:
Assay buffer: PBS 0.05% Tween20 with 0.1% (w/v) BSA
19.4 ml PBS
200 µl 10% Tween20 in water or PBS
400 µl 5% BSA
Freshly made in RT
COMMENTARY:
Background information
The basic protocol is a robust method with limitations. One limitation is that the affinity and stability of the humanized VNARs can be different from the original, and the grafted sequences may not cover all the antigen binding residues (ABR). In our experience with rabbit and mouse antibody humanization (Zhang et al., 2016, 2017; Zhang et al., 2015), it is normal to see affinity increase (e.g., hYP218) in ~1/4 of cases, and affinity decrease <3 fold sometimes. In Table 1, we list all the antigen-contacting residues in Figure 2. There are some positions outside of the grafted regions in the basic protocol, and these positions can be grafted in subsequent sequences for further optimization if necessary. Such work may lead to future improvement in the humanization technology.
Table 1.
Positions of ABR and grafted sequences in the basic protocol.
| Region | KABAT position according to aligned human DPK9_JK1 sequence |
|---|---|
| FW1 | 2, 4, 5, 7, 22 |
| CDR1 | 24, 25, 26, 27, 27a, 27b, 28, 29, 31, 32, 33, 34 |
| FW2 | 36, 38, 39 |
| HV2 | 43, 44, 45, 46, 47, 51 |
| FW3a | 52, 55, 57, 58, 65 |
| HV4 | 66, 68, 69, 70 |
| FW3b | 72, 85, 87 |
| CDR3 | 89~100 |
| FW4 | 101, 103 |
Red: Grafted in the basic protocol
In particular:
The N-terminus of the FW 1 region frequently contacts the antigen in shark VNARs as they do in mammalian antibodies (Zhang et al., 2017), and the N-terminal ABR in VNARs can extend to position 7. Therefore position 7 can be grafted during sequence optimization if needed.
The FW 2 position 36 is not grafted in the basic protocol, because in this example the shark VNAR and human DPK9_JK1 sequence are both Y (tyrosine). But in some cases, the shark VNAR KABAT position 36 is F (Phenylalanine), and the 36F can contact the antigen (Figure 2A). Therefore, it is advisable to graft position 36 when it differs from the human sequence.
We found the shark VNAR CDR3 often has a longer C-terminus than the mammalian counterparts, so we graft positions 89 through 100, but in some VNARs the ABR extends through 103. Therefore, we can graft residues 89 through 103 during sequence optimization if needed.
Steven et al. described a way to further improve the humanized sequence by generating and panning on a mutation library in phage (Steven et al., 2017), and this is also a valid approach to consider. Briefly, the lead humanized sequence (e.g. the humanized sequence from the basic protocol) is mutagenized by error-prone PCR, and then cloned into a phage display vector to make a library of plasmids. The plasmid library can then be transformed into TG1 cells to make a phage library, and then the library is panned to select the desired binders. From construction of the mutant phage library to completion of the panning can take months and need advanced antibody engineer skills.
Another human template that can be considered besides DPK9 is DPK24 (Steven et al., 2017). In the case of E06, DPK9 based humanized E06 tend to dimerize (partly due to its CDR3 sequence), whereas DPK24 based humanized E06 is monomeric and expressed well, but had much lower affinity than E06. After affinity maturation (Steven et al., 2017), both templates can give monomeric proteins with higher affinity than the original humanized versions.
Critical Parameters:
Unpaired cysteine naturally appears in antibodies of all species and is often undesirable for future development of therapeutic antibodies. If the unpaired cysteine is in the grafted portion of VNAR, one can generate a second humanized sequence by mutating the unpaired cysteine to serine. If the unpaired cysteine is not in the grafted portion of VNAR, after CDR grafting the unpaired cysteine will be removed because the human scaffold has no unpaired cysteine.
In kinetic measurement experiment using Octet, the VNAR loading concentration should be predetermined to avoid over-crowding the octet probe(Sultana et al., 2015). The detailed protocol can be found in octet application note page 12–13: optimizing ligand loading density (https://www.sartorius.com/resource/blob/742330/05671fe2de45d16bd72b8078ac28980d/octet-biomolecular-binding-kinetics-application-note-4014-en-1--data.pdf). In the case of B6 and hB6, we used 5 µg/ml as loading concentration.
Troubleshooting: Table 2
Table 2.
Troubleshooting
| Problem | Possible cause | Solution |
|---|---|---|
| VNAR sequence does not align with DPK9-JK1 sequence very well | VNAR sequence is too different from human | Try inputting additional shark VNAR sequences provided in this protocol in figures 2 or 3 into Clustal omega, so that other VNAR sequences can guide the alignment. Choose the additional shark VNAR sequences based on sequence identity and the four types of VNAR. Look for canonical sequences to help guide the alignment. Sometimes there can be additional unpaired cysteines in the sequence, and if they are not at the canonical locations, they should not be aligned with the canonical cysteines in other VNARs. |
| Humanized VNAR does not bind or has greatly reduced binding | Wrong alignment, antigen-contacting residues changed | Make sure the alignment is correct by visually checking the alignment. Mutate the humanized sequence back to the shark sequence site by site. Use Figure 2 in this protocol to check potential back-mutation sites. Pay special attention to CDR-framework borders and HV-framework borders as border residues often play an important role in folding. |
| Non-specific binding in octet | Tween20 and BSA concentrations are not optimal | Try different Tween20 (up to 0.09%) and BSA (up to 1~2%) concentrations in assay buffer. Always pre hydrate the probe for at least 10 minutes. Use high quality proteins such as size exclusion column purified proteins. |
Understanding Results:
Although the binding affinity is summarized as KD (and Bmax, Kon and Koff), there are other factors to consider in the evaluation of the humanization, such as immunogenicity, stability, expression level, cell binding, bioactivity, etc. More human sequences in humanized antibodies does not always correlate with lower affinity or stability (Zhang et al., 2017; Zhang et al., 2015). We have developed humanized mouse (D. Li et al., 2020; Zhang et al., 2016) and rabbit (Tomar et al., 2022; Zhang et al., 2015) antibodies into clinical stage therapeutics. To this end, we have tested humanized antibodies in various clinical formats such as chimeric antigen receptor (CAR)-T cells (D. Li et al., 2020; Tomar et al., 2022), antibody-drug conjugates (ADCs)(Fu et al., 2019) and T-cell engager bispecific antibodies (Chen et al., 2022).
Time Considerations: Table 3
Table 3.
Time considerations
| Procedure | Time |
|---|---|
| Design of the first candidate sequence using the basic protocol | 1–2 h |
| Use Alphafold to generate structure models | 1 day |
| Use Dali server to align the structure models, analyze superimposed structure models | Half day |
| Clone (h)VNAR into expression vector | 1 week |
| Make and purify the protein in E. coli | 3–5 days |
| Optional: Make the protein in Expi293F cells (not including cloning) | ~2 weeks |
|
| |
| Affinity measurement using octet | 1–2 days |
Supplementary Material
ACKNOWLEDGEMENTS:
This project was supported by the Intramural Research Program of the NIH, Center for Cancer Research (CCR), National Cancer Institute (NCI) (Z01 BC010891 and ZIA BC010891 to M.H.) and by the NCI CCR Antibody Engineering Program (ZIC BC 011891 to M.H.). We thank Mingqian Feng, a former member in our laboratory (NCI), for isolating B6 VNAR by phage display technology, Gregory Piszczek and Di Wu from the Biophysics Core Facility in National Heart, Lung, and Blood Institute for Octet assay technical support. We also thank NIH Fellows Editorial Board for editorial assistance. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
CONFLICT OF INTEREST STATEMENT:
The authors declare no conflict of interest.
DATA AVAILABILITY STATEMENT:
The data, tools, and materials (or their source) that support the protocol are available from the corresponding author upon reasonable request.
Literature cited
- Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, … Shepard HM (1992). Humanization of an anti-p185HER2 antibody for human cancer therapy. Proceedings of the National Academy of Sciences of the United States of America, 89(10), 4285–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chen Y, Liang R, Xiang L, Li J, Zhu Y, … Feng M (2022). Combination Therapy of Hepatocellular Carcinoma by GPC3-Targeted Bispecific Antibody and Irinotecan is Potent in Suppressing Tumor Growth in Mice. Molecular Cancer Therapeutics, 21(1), 149–158. doi: 10.1158/1535-7163.mct-20-1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C, & Lesk AM (1987). Canonical structures for the hypervariable regions of immunoglobulins. Journal of Molecular Biology, 196(4), 901–917. doi: 10.1016/0022-2836(87)90412-8 [DOI] [PubMed] [Google Scholar]
- Dooley H, Stanfield RL, Brady RA, & Flajnik MF (2006). First molecular and biochemical analysis of in vivo affinity maturation in an ectothermic vertebrate. Proceedings of the National Academy of Sciences of the United States of America, 103(6), 1846–1851. doi: 10.1073/pnas.0508341103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Buffington J, Hong J, & Ho M (2022). Production and Purification of Shark and Camel Single-Domain Antibodies from Bacterial and Mammalian Cell Expression Systems. Curr Protoc, 2(6), e459. doi: 10.1002/cpz1.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Buffington J, Hong J, & Ho M (2022). Production and Purification of Shark and Camel Single-Domain Antibodies from Bacterial and Mammalian Cell Expression Systems. Curr Protoc, 2(6), e459. doi: 10.1002/cpz1.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English H, Hong J, & Ho M (2020a). Ancient species offers contemporary therapeutics: an update on shark V(NAR) single domain antibody sequences, phage libraries and potential clinical applications. Antib Ther, 3(1), 1–9. doi: 10.1093/abt/tbaa001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English H, Hong J, & Ho M (2020b). Ancient species offers contemporary therapeutics: an update on shark VNAR single domain antibody sequences, phage libraries and potential clinical applications. Antib Ther doi: 10.1093/abt/tbaa001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng M, Bian H, Wu X, Fu T, Fu Y, Hong J, … Ho M (2019). Construction and next-generation sequencing analysis of a large phage-displayed V(NAR) single-domain antibody library from six naïve nurse sharks. Antib Ther, 2(1), 1–11. doi: 10.1093/abt/tby011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell BJ, Darmanin-Sheehan A, Hufton SE, Calabro V, Wu L, Müller MR, … Finlay WJ. J. (2010). Dissection of the IgNAR V Domain: Molecular Scanning and Orthologue Database Mining Define Novel IgNAR Hallmarks and Affinity Maturation Mechanisms. Journal of Molecular Biology, 400(2), 155–170. doi: 10.1016/j.jmb.2010.04.061 [DOI] [PubMed] [Google Scholar]
- Flajnik MF (2018). A cold-blooded view of adaptive immunity. Nature Reviews: Immunology, 18(7), 438–453. doi: 10.1038/s41577-018-0003-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Urban DJ, Nani RR, Zhang Y-F, Li N, Fu H, … Ho M (2019). Glypican-3-Specific Antibody Drug Conjugates Targeting Hepatocellular Carcinoma. Hepatology, 70(2), 563–576. doi: 10.1002/hep.30326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, & Flajnik MF (1995). A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature, 374(6518), 168–173. doi: 10.1038/374168a0 [DOI] [PubMed] [Google Scholar]
- Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, … Hamers R (1993). Naturally occurring antibodies devoid of light chains. Nature, 363(6428), 446–448. doi: 10.1038/363446a0 [DOI] [PubMed] [Google Scholar]
- Holm L (2020). Using Dali for Protein Structure Comparison. In Gáspári Z (Ed.), Structural Bioinformatics: Methods and Protocols (pp. 29–42). New York, NY: Springer US. [DOI] [PubMed] [Google Scholar]
- Jones PT, Dear PH, Foote J, Neuberger MS, & Winter G (1986). Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature, 321(6069), 522–525. [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, … Hassabis D(2021). Highly accurate protein structure prediction with AlphaFold. Nature, 596(7873), 583–589. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat EA, Te Wu T, Perry HM, Gottesman KS, & Foeller C (1992). Sequences of Proteins of Immunological Interest: Diane Publishing Company. [Google Scholar]
- Kovalenko OV, Olland A, Piché-Nicholas N, Godbole A, King D, Svenson K, … Tchistiakova L (2013). Atypical Antigen Recognition Mode of a Shark Immunoglobulin New Antigen Receptor (IgNAR) Variable Domain Characterized by Humanization and Structural Analysis. Journal of Biological Chemistry, 288(24), 17408–17419. doi: 10.1074/jbc.M112.435289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunik V, Peters B, & Ofran Y (2012). Structural Consensus among Antibodies Defines the Antigen Binding Site. PLoS Computational Biology, 8(2), e1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Li N, Zhang Y-F, Fu H, Feng M, Schneider D, … Ho M (2020). Persistent Polyfunctional Chimeric Antigen Receptor T Cells That Target Glypican 3 Eliminate Orthotopic Hepatocellular Carcinomas in Mice. Gastroenterology, 158(8), 2250–2265.e2220. doi: 10.1053/j.gastro.2020.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Fu H, Hewitt SM, Dimitrov DS, & Ho M (2017). Therapeutically targeting glypican-2 via single-domain antibody-based chimeric antigen receptors and immunotoxins in neuroblastoma. Proceedings of the National Academy of Sciences, 114(32), E6623–E6631. doi: 10.1073/pnas.1706055114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padlan EA, Abergel C, & Tipper JP (1995). Identification of specificity-determining residues in antibodies. The FASEB Journal, 9(1), 133–139. [DOI] [PubMed] [Google Scholar]
- Pastan I, & Ho M (2010). Recombinant immunotoxins for treating cancer. In Kontermann R & Dubel S (Eds.), Antibody Engineering (2 ed., Vol. 2, pp. 127–146). Berlin-Heidelberg: Springer-Verlag. [Google Scholar]
- Queen C, Schneider WP, Selick HE, Payne PW, Landolfi NF, Duncan JF, … Waldmann TA (1989). A humanized antibody that binds to the interleukin 2 receptor. Proceedings of the National Academy of Sciences of the United States of America, 86(24), 10029–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield RL, Dooley H, Flajnik MF, & Wilson IA (2004). Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science, 305(5691), 1770–1773. doi: 10.1126/science.1101148 [DOI] [PubMed] [Google Scholar]
- Steven J, Müller MR, Carvalho MF, Ubah OC, Kovaleva M, Donohoe G, … Barelle CJ (2017). In Vitro Maturation of a Humanized Shark VNAR Domain to Improve Its Biophysical Properties to Facilitate Clinical Development. Frontiers in Immunology, 8. doi: 10.3389/fimmu.2017.01361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streltsov VA, Varghese JN, Carmichael JA, Irving RA, Hudson PJ, & Nuttall SD (2004). Structural evidence for evolution of shark Ig new antigen receptor variable domain antibodies from a cell-surface receptor. Proceedings of the National Academy of Sciences, 101(34), 12444–12449. doi:doi: 10.1073/pnas.0403509101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana A, & Lee JE (2015). Measuring Protein-Protein and Protein-Nucleic Acid Interactions by Biolayer Interferometry. Current Protocols in Protein Science, 79(1), 19.25.11–19.25.26. doi: 10.1002/0471140864.ps1925s79 [DOI] [PubMed] [Google Scholar]
- Tomar S, Zhang J, Khanal M, Hong J, Venugopalan A, Jiang Q, … Hassan R (2022). Development of Highly Effective Anti-Mesothelin hYP218 Chimeric Antigen Receptor T Cells With Increased Tumor Infiltration and Persistence for Treating Solid Tumors. Molecular Cancer Therapeutics, 21(7), 1195–1206. doi: 10.1158/1535-7163.mct-22-0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Žídek A, … Hassabis D (2021). Highly accurate protein structure prediction for the human proteome. Nature, 596(7873), 590–596. doi: 10.1038/s41586-021-03828-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, … Wilson KS (2011). Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography, 67(4), 235–242. doi:doi: 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, & Kabat EA (1970). An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. The Journal of Experimental Medicine, 132(2), 211–250. doi: 10.1084/jem.132.2.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-F, & Ho M (2016). Humanization of high-affinity antibodies targeting glypican-3 in hepatocellular carcinoma. Scientific Reports, 6, 33878. doi: 10.1038/srep33878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-F, & Ho M (2017). Humanization of rabbit monoclonal antibodies via grafting combined Kabat/IMGT/Paratome complementarity-determining regions: rationale and examples. mAbs, 00–00. doi: 10.1080/19420862.2017.1289302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-F, Phung Y, Gao W, Kawa S, Hassan R, Pastan I, & Ho M (2015). New High Affinity Monoclonal Antibodies Recognize Non-Overlapping Epitopes On Mesothelin For Monitoring And Treating Mesothelioma. Scientific Reports, 5, 9928. doi:10.1038/srep09928 http://www.nature.com/articles/srep09928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielonka S, Empting M, Grzeschik J, Könning D, Barelle CJ, & Kolmar H (2015). Structural insights and biomedical potential of IgNAR scaffolds from sharks. mAbs, 7(1), 15–25. doi: 10.4161/19420862.2015.989032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data, tools, and materials (or their source) that support the protocol are available from the corresponding author upon reasonable request.


