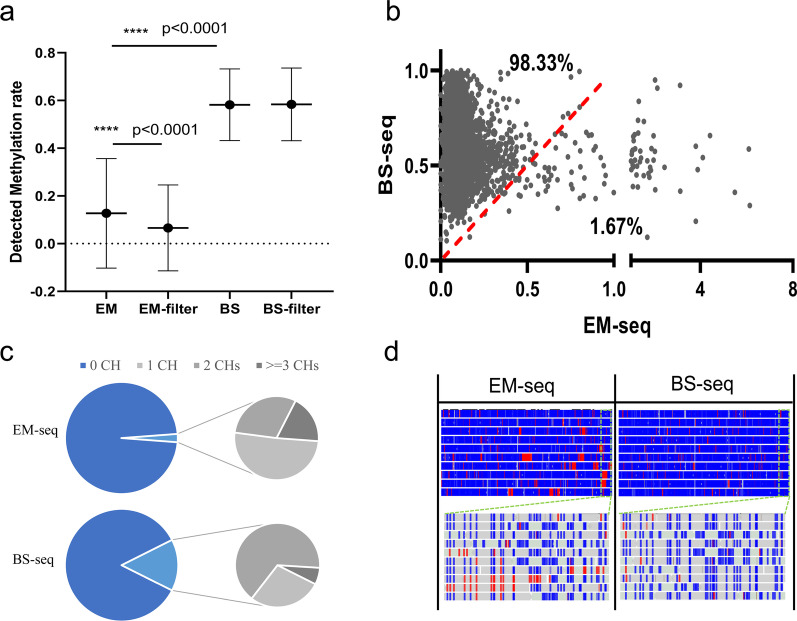
Fig. 1.
In complete conversion in EM-seq. a Methylation values on hypomethylated CpGs acquired by EM-Seq and BS-seq, before and after ≥ 3CHs filtration. Hypomethylated CpGs are those with BS-seq detection values of < 1% on λ DNA. b Dot plot compares individual methylation values in hypomethylated CpGs acquired by EM-Seq and BS-seq. Percentages indicate the fraction of CpGs that differed between conditions. c Pie charts compare the proportion of reads with 0, 1, 2, and ≥ 3 CH sites in all EM-seq and BS-seq sequencing reads. d. Genome plot for unmethylated control λ DNA (pos: 30,000–35,000) compares CH reads between EM-seq and BS-seq datasets. Boxes represent reads, and unmethylated (blue) and methylated (red) CHHs are indicated