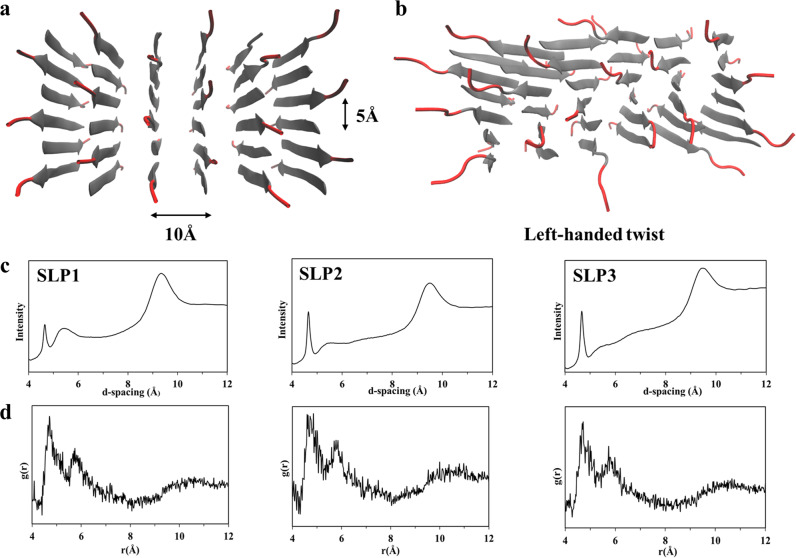
Fig. 5. Fibril models derived from MD simulations.
a Representative atomistic manually built model of the starting fibril configuration with antiparallel SLPs. SLPs within a β-sheet are 5 Å apart, and 10 Å is the orthogonal distance between SLPs. b Representative final configuration of the equilibrated molecular models after 100 ns of MD simulation. The chirality of the L-amino acids leads to left-handed twist as is observed in the SLP models. c XRD pattern of the SLP1–3 fibrils. The reflection at ~4.7 and 9–11 Å represent the signatory of cross-β diffraction pattern. d Radial distribution function (RDF) of the backbone–backbone distance calculated from the final MD configuration of SLP fibrils. In agreement with XRD, RDF also show peaks at ~4.7 and 9–11 Å, demonstrating a good match between MD fibril models and experimental data.