Summary
Hundreds of studies in last decades have aimed to compare the microbiome of patients suffering from diverse diseases with that of healthy controls. The microbiome-related component was additionally identified in pathophysiology of many diseases formerly considered to depend only on the host physiology. This, however, opens important questions like: “What is the healthy microbiome?” or “Is it possible to define it unequivocally?”. In this review, we describe the main hindrances complicating the definition of “healthy microbiome” in terms of microbiota composition. We discuss the human microbiome from the perspective of classical ecology and we advocate for the shift from the stress on microbiota composition to the functions that microbiome ensures for the host. Finally, we propose to leave the concept of ideal healthy microbiome and replace it by focus on microbiome advantageous for the host, which always depends on the specific context like the age, genetics, dietary habits, body site or physiological state.
Keywords: Holobiont, Core microbiome function, Resilience, Microbiome ecology, One health hypothesis, Dysbiosis
Holobiont concept
Progress in sequencing techniques opened new areas of research and revealed that all multicellular organisms, including humans, live in a tight co-existence with rich and highly variable resident microbiota (the bacteria, archaea, viruses, and fungi) that have a significant influence on the host development and health. The term microbiota is, in the literature, often replaced by the term microbiome, which, however, has two other, equally used, distinct definitions. The genetic definition uses this term to describe only the sum of genetic information of the resident microbiota, while the ecological point of view uses the term to describe the microbial community including its habitat with typical physical and chemical conditions, i.e. a dynamic and interactive microecosystem. Additionally, when speaking about microbiota or microbiome, the researchers often have in mind only bacteria, as they represent the most abundant and also most studied portion of microbial population associated with human body. This is also the case of this review.
The human associated microbiota (microbiome) is now being recognized as a “new organ” that complements the host’s missing functions. Research focused on the role of microbiota in health and disease or on microbiome-based therapy, opens questions like: “What are the most important characteristics of healthy microbiome?”, “What core functions should it ensure for the host?” and “How could it be described?”. Growing understanding of the complexity of microbial ecosystems and their relationships with their environment unravels that there is probably nothing like one ideal healthy microbiome community.
The achievements in the study of the human microbiome shifted the perception of multicellular organisms: they are not only a single entity by themselves, but should be considered as a whole together with a highly variable resident microbiota (the bacteria, archaea, viruses, and fungi), hence the term “holobionts” [1]. Both the eukaryotic and prokaryotic components are tightly interconnected and live in a state of dynamic balance. Furthermore, the microbiome component is being continually challenged and replenished by contact with the surrounding environment (Fig. 1).
Fig. 1.
Human holobiont and its interaction with the environment. © Linda Čihařová.
The holobiont concept brought yet another new term, hologenome, describing collective genomes of the host and its microbiota, where the host (human, animal, plant etc.) genes are only a minority. In the human holobiont, microbial genomes probably outnumber the human genes approx. 100times [2]. From this perspective, even a birth event is not only a new human, but also a new community “infant plus its microbiota” [3]. Since the start of the Human Microbiome Project, scientists have aimed to characterize the human healthy/beneficial microbiota, however, even after more than a decade there is still no sufficient insight on its nature or how it should behave. Here we summarize the main challenges we face in the field and highlight the most promising approaches.
Human-associated microbiome from the ecological perspective
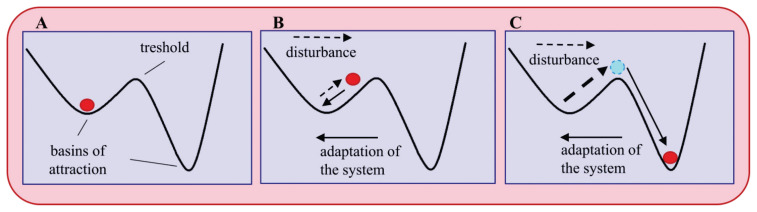
Microbial communities inhabiting various niches of the human body are communities that meet the criteria of macroecosystems and therefore, it is useful and justifiable to borrow the concepts and methods from classical ecology. The ecosystem consists of all organisms living in a defined area and their interaction with the physical environment. This definition encompasses the complex, adaptive system that is characterized by historical dependency, nonlinear dynamics, threshold effects (i.e. factors promoting the return to the stable state after the disturbance), multiple basins of attraction (i.e. stable states), and limited predictability [4].
The behavior of the system could be described using the model of “stability landscape” [5]. In this model, the basins of attraction [depressions] represent the stable states. Within the basins, the systems tend to return to equilibrium with the lowest energy. The disturbances, i.e. substantial changes in the environment or community structure and composition, allow the system to pass the threshold and to set in a new stable state (Fig. 2). They are invaluable sources of stimuli leading to ecosystem adaptation and evolution if they occur in a predictive manner and manageable scale. On the other hand, if it is unpredictable and erratic, the community would suffer losses and even eventually become extinct [6].
Fig. 2.
Stability landscape model. (A) original state; (B) transition state; (C) new stable state. Dashed arrows indicates the disturbance, solid arrows the adaptation of the system to the disturbance. Adapted according to Folke et al. [5].
Stability, resistance, and resilience are essential characteristics of any ecosystem including the human microbiome [7]. According to Pimm, a system is stable if key variables describing the system return to equilibrium values after displacement, the functions of the system are maintained and there is limited variability of key system parameters over time [8]. Resistance is defined as the capacity of an ecosystem to remain unchanged on perturbation [9]. Ecological resilience was conceptualized by Holing in 1996 and could be defined as a capacity of a system to absorb disturbance and reorganize while undergoing change, so as to still retain essentially the same function, structure, identity, and feedback [10]. The combined and often synergistic effects of anthropogenic pressures can make ecosystems less resilient and thus more vulnerable to changes that could have been previously absorbed.
In the human microbiome context, the initial state (Fig. 2A) may represent the stable microbiome of a healthy individual [i.e. advantageous for the host]. An intermediate level of disturbance modifies the community composition and its metabolic function, but the microbiome can revert to the original state (Fig. 2B). If the intensity of the disturbance exceeds the adaptive capacity of the ecosystem, it passes the threshold and reaches a new stable state (Fig. 2C). The mild disturbance might be a diversified diet or an exposure to a microbial-rich environment. An intensive disturbance could be provoked by the massive use of antibiotics, extensive sanitation, etc. and will push the system to a new stable state, potentially disadvantageous for the host (further referred as unhealthy or dysbiotic).
The stable microbial system is intuitively considered healthy and indeed, it probably is – but from its own point of view, i.e. from point of view of the microbiome – not necessarily also the host. For example, in inflammatory bowel disease [IBD] or recurrent Clostridioides difficile infection, the gut microbiome could also be stable and resilient and as such, it becomes a significant obstacle to therapeutic intervention and contributes to the chronicity of the disease [11,12]. A stable state per se is not a sufficient indicator of a beneficial function, but understanding how stability is established and maintained is essential for diagnosis and successful therapy of many diseases.
A key factor for microbiome stability and resilience is the microbial diversity and the consequent functional redundancy. This observation, originally described in grassland savanna ecosystems [13], was repeated in a laboratory “micro-setting”. Naeem and Li performed an experiment on a wide set of artificial microbial communities with a different representation of key functional microbial groups representing terrestrial and aquatic ecosystems and variable amount of available nutrients. They found that the capacity of the system to maintain productivity was dependent on the balanced representation of the number of species per functional group and concluded that the “redundancy is a valuable commodity” [14].
These observations resulted in the formulation of the “biological insurance hypothesis” [15] according to which compensation by one species for loss or decline in another preserves long-term average ecosystem performance and reduces variability in performance, promotes the long-term probability of persistence, and enhances resilience to perturbations.
How to define “being healthy”?
Even though the question seems to be simple, the answer is extremely complicated. The first problem represents the term “healthy”. Oxford Dictionary defines health as “the state of being free from illness and injury”. On the opposite end of the scale is WHO definition that describes health as “a state of complete physical, mental and social wellbeing and not merely the absence of disease or infirmity”. Both definitions received substantial criticism. While the former is negative and only excludes the state of illness, the latter is too complex and impossible to measure. Furthermore, the increase in the prevalence of chronic diseases would mean that many people with even minor health complications would be persistently considered as being ill [16, 17]. Despite the profound differences between these two definitions, they both share one common feature – they are static.
In 1982, Stokes et al. proposed following definition: “Health is a state characterized by anatomical, physiological, and psychological integrity; an ability to perform personally valued family, work, and community roles; an ability to deal with physical, biological, psychological, and social stress.” [18]. Interestingly, this definition introduces an important aspect, the ability to cope with stress, which moves the perception of health towards a dynamics process – seeking a balance. From this perspective, health is “a dynamic condition, encompassing resilience to stress and recovery from damage” [16,17].
Human microbiome[s] are very dynamic structures and there is no way to define and describe if they are a priori beneficial or harmful. The concept of dynamic health allows characterizing healthy or pathological microbiomes according to specific conditions. According to the concept of the human holobiont, illness could be related to an non-resilient microbiome unable to meet the physiological demands of the host [19].
Healthy microbiome in the “one health concept”
According to one health concept in its simplified version, it is impossible or at least highly improbable to stay healthy in an unhealthy environment. To understand the holobiont physiology in its complexity we should, therefore, consider not only the two-component system, i.e. the host and its microbiome, and their mutual interactions, but also the holobiont interactions with its environment.
The maintenance of a healthy microbiome is critically dependent on the continuous acquisition of microorganisms and appropriate supporting substrates through feeding, drinking, breathing, and other interactions with the environment [20–22]. For example, the gut microbiome of hunters and gatherers still surviving in small communities living in relatively pristine areas and in close contact with their natural surroundings is characterized by a higher stability as well as higher diversity when compared to the western population living in urban areas. The gut microbiome diversity of western populations is reduced at all taxonomic levels, meaning that not only species, but also whole large groups, encompassing hundreds of species, are absent. This results in a loss of redundancy and thus of essential functions. The diet common in western societies, characterized by an oversupply of animal protein and fat and a low amount of plant polysaccharides, is associated with a poor capacity to digest carbohydrates [23, 24].
Paradoxically, recent advances in medicine and better housing act against the natural self-renewing capacity of our microbiome resulting from the close exposure to the external reservoirs. The massive exposure to antibiotics often results in the depletion of keystone bacterial taxa or whole functional groups called guilds [22,25,26]. Resulting changes in microbiome composition of citizens of developed countries have been correlated with a low level of resilience, chronic sub-inflammation, and compromised setting of the immune system [27, 28]. The “hygiene hypothesis” postulates that reduction in the frequency of infections contributes directly to the increase in the frequency of autoimmune and allergic diseases while the contact with environments rich in microbial diversity protects against these disorders [29–31].
Microbiome-related diseases
The enormously growing microbiome research has important implications in the perception of the mechanisms underlying the onset and development of many NCDs. The way of life in modern, westernized society is profoundly different from the conditions determining the co-evolution of human hosts and their microbiomes. Relatively mild, but long-term influence of conditions like western-type lifestyle with unhealthy diets [32–34], high hygiene standards and extensive usage of cosmetics [35–37], overuse of medicine including antibiotics and proton-pump inhibitors [38], disturbances of the circadian rhythm [39,40] etc. can cause the deterioration of the human body-associated microbiome ecosystem. In detail, it could be manifested as the loss of key bacterial taxa/guilds, loss or reduction of essential microbiome-mediated functions and metabolites, aberrant stimulation of immune system and compromised control against pathogen attack [19,22,41]. Such changes may belong to the principal drivers of the rise of non-communicable diseases (NCDs) prevalence throughout the last decades [27,42,43]. The traditional definition of NCDs like asthma, heart disease, obesity, type 2 diabetes, cancer, neurodegenerative conditions or autoimmune diseases rules out microbes as causative agents. Recently the links between NCDs and altered, mainly, but not exclusively, gut microbiome were reported and the therapeutic implications have attracted keen interest among scientists. Several studies suggest that at least in some NCDs there is substantial microbiota-related component and thus they may be to some degree communicable among humans [43, 44]. That might as well be the case, but some caution when interpreting the data and translating them into human context is desirable. Many of the disease – microbiota associations are based on correlation studies, i.e. comparison of microbiota in apparently healthy and diseased population. This type of study suffer from two limitations, i.e. (i) correlation does not prove causation [45] and (ii) symptomatically invisible dysbiosis often precedes the disease onset as will be discussed further. The widely used proof-of-concept approach is the transplantation of fecal microbiota from individuals with and without a disease into germ-free animals. The subsequent recapitulation of the diseased phenotype is considered as the proof of causality and was demonstrated for many pathophysiological states, e.g. cardiovascular disease[46], IBD [47], type 2 diabetes [48], obesity [49] and others. Even though the outcomes of these studies are generally accepted, this experimental design has inherent limitations complicating the interpretation of the results [50]. The authors definitely do not intend taking the role of microbiome in health and disease into question but it is necessary to keep in mind that oversimplified associations may lead to misinterpretation of experimental results and false identification of specific microbiota composition as “healthy” or “dysbiotic”.
Microbiome-focused therapy
Having in mind the holobiont concept, it seems shortsighted to focus the medical and scientific attention only on the host and his/her physiological processes and to neglect the therapeutic potential of our co-inhabitants. The identification of microbiota-related component in various diseases opens new field of microbiome-focused therapy that may be either untargeted [probiotics, prebiotics, fecal microbiota transfer] or targeted [engineered bacteria, postbiotics, phages] [51]. Among these options, the fecal microbiota transfer (FMT) has the greatest potential to induce significant shift in whole gut microbiota community [12] and therefore, to replenish the missing function[s] of the microbiome in complexity. Currently, only recurrent Clostridioides difficile infection is approved for FMT therapy in both USA and EU [52] but at this moment, there are 150 clinical trials registered in clinicaltrials.gov investigating FMT therapeutic potential in many other pathological conditions, i.e. IBD, obesity, liver diseases, neurological diseases etc. One of the main challenges that hinder wider application of this otherwise safe and inexpensive therapy in clinical practice is the lack of reliable criteria for the donor. According to the current standards, donors are meticulously tested for potential pathogen presence but the risk of the transmission of more complex “microbiota setting” is still not addressed. Indeed, one case study documented the transmission of an obese phenotype from an overweight donor to a lean patient following FMT for Clostridioides difficile infection [52,53]. Particularly from this point of view, the definition of healthy microbiome is of utmost importance. In the following paragraphs, we will discuss the uniqueness of host-microbiome interaction what opens the question whether it is feasible to establish the requirements of one super-donor or whether is better to adjust the requirements for the donor to the needs of specific recipient.
There is nothing like “human healthy microbiome”
It seems a healthy microbiome ensures a better health. However, the fundamental question, how the healthy microbiome should look like, has not been answered. To describe the healthy microbiome, we face several challenges, related to its variability in both time and space: 1) the individual microbiome exhibits both long-term and short-term dynamics. 2) Each body niche harbor a different microbial community adapted to highly variable local conditions. 3) The microbiota communities of different body niches are not separated but interact and influence each other. Therefore, the “unhealthy” state originating in one location may spread to other niches. 4) Since the dysbiosis often precedes the clinical signs of the disease, the microbiome of an apparently healthy individual can already be dysbiotic. 5) Usefulness of specific microbiota for the host is context-dependent. Specific microbiota can, depending on other circumstances, represent both the life-saving condition as well as the serious threat.
Dynamic character of the microbiome
The short-term fluctuations are caused for example by a change in the type of the physiological status, circadian rhythms, mechanical stimuli etc. while the long-term variability can result from hormonal shifts or changes connected with aging. The oral microbiome undergoes daily short-term dynamics. The tooth surface and supragingival community is challenged several times a day by teeth-brushing or intake of some foodstuff [e.g. simple sugars] and is naturally restored from other niches in the mouth as well as from the external sources [54]. The vaginal microbiome in some women exhibited remarkable variations in time during the menstrual cycle, however for other women it remained relatively stable [55]. The skin microbiome is generally considered to be highly stable in time, however, some parts of the foot also exhibited remarkable variability [56].
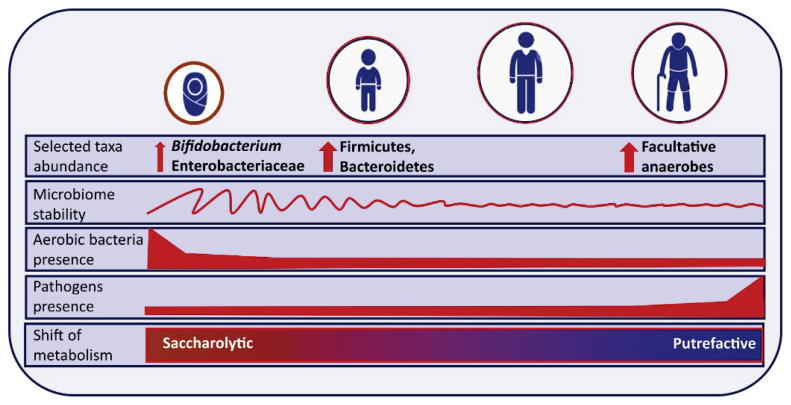
The long-term dynamics of the human microbiome are driven by physiological changes related to ontogenesis and aging (Fig. 3). In this context, the gut microbiome is probably the most studied one. During the very first days/weeks, the newborn gut microbiome is dominated by aerobic and facultative anaerobic bacteria. As the oxygen content in the gut gradually decreases, obligate anaerobes subsequently prevail. By the age of three years, the distal gut microbiota composition is represented almost entirely by obligate anaerobes [57]. After the third year, the gut microbiome becomes less dynamic, however, the stable adult microbiome is established approximately at the end of the second decade of life and persists, again only approximately, up to the age of seventy. Aged microbiome is characterized by a continuous decline in the physiological functions affecting a wide spectrum of metabolic and immunological processes [58] resulting in a chronic pro-inflammatory status called “inflammaging”. Despite significant individual and geographical variability, there are some common features of age-related changes in gut microbiota composition: (i) decreased alpha diversity [59]; (ii) increase of potentially pathogenic bacteria, e.g. Streptococcaceae, Staphylococcaceae, and Enterobac-teriaceae [60]; (iii) reduction of the abundance of potentially beneficial bacteria like Faecalibacterium prausnitzii, Roseburia or Bifidobacterium [27,61]. Finally, the changes in microbiota composition are reflected by an altered functional performance, e.g. decreased production of beneficial short-chain fatty acids and increased production of branched-chain fatty acids. In general, aging is associated with a shift from predominantly saccharolytic metabolism towards predominantly putrefactive metabolism in the elderly, with more fermentation of proteins, which concomitantly produces different harmful fermentation metabolites [62,63]
Fig. 3.
Long-life dynamics of gut microbiome.
The variability of human body niches
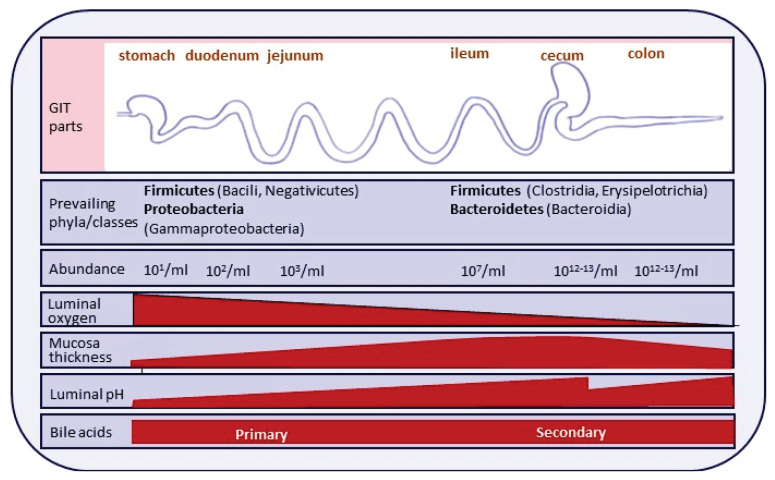
The multicellular organism is composed of many, often highly variable, niches providing its microbial inhabitants with a wide range of living conditions. The oxygen pressure varies from fully aerobic; e.g. on the skin, to strictly anaerobic conditions; e.g. in the deep periodontal pockets or in the distal gut [cecum and colon]. The temperature may be quite stable (~37 °C) in the gastrointestinal (GIT) or urogenital tract or highly variable on the skin surface depending on the environment, activities, and living habits of the host. The pH value can vary from strongly acidic in the stomach (pH=2), mildly acidic on the skin surface [pH=5.5] to more-less neutral in the oral cavity or small intestine. The energy sources vary a lot even throughout the gastrointestinal tract (GIT) and of course, any part of the GIT will provide a more rich and variable source of energy when compared for example to the vagina or scalp.
GIT harbors many extremely different microbial communities. In the healthy oral cavity, there are at least four diverse ecological niches: the tongue, buccal mucosa, teeth surface, and gingival crevice, which differ in oxygen and nutrition availability, and saliva flow. In fact, the saliva could be considered another ecological niche. Continuing further through the GIT, the dominant environmental factors affecting the microbiome composition are acidity, oxygen pressure, bile acid composition and nutrient availability (Fig. 4).
Fig. 4.
Variability of the environmental conditions along the gastrointestinal tract.
The body surface provides variable environments as well. In general, we distinguish dry, moist and oily (sebaceous) areas on the skin and in addition some areas exhibiting topography-related specific features (foot toes), each harboring distinct microbial communities, for review see [56]. The oily sites are typically colonized by Cutibacterium species while the moist environment of groin or navel is more suitable for Corynebacteriaceae and the bottom of heal is dominated by Staphylococcaceae [64]. This skin microbiome variability is, however, true rather for the skin surface. Bay et al. demonstrated, that the microbiome of the lower dermal layers exhibits lower topologic diversity, is well conserved, and functionally distinct from the epidermal community [65].
Interaction of individual microbiomes/body niches
The microbiota communities separated in space communicate both directly, e.g. by transfer of microbiota and other material including microbial metabolites through the GIT, and indirectly via influencing immune system, neural network, and/or hormones. The most studied model is the oral-gut axis. On average, humans swallow 1.5 liters of saliva containing 1.5 × 1012 bacteria per day [66, 67]. The oral and gut microbiome seemed to be separated by physical barriers and chemical hurdles like a strongly acidic milieu in the stomach or primary bile acids in the duodenum. Despite these obstacles, however, the presence of oral bacteria has been demonstrated in many body sites [68–72]. Live oral bacteria were described not only in lower GIT, but also in the aortic tissue [73], skin [74,75], atherosclerotic plaques [76], human breast milk [77], brain of Alzheimer-affected patients [78], and healthy placenta [79]. For long, the translocation of oral bacteria into lower GIT and other locations was considered to be rare, and it was supposed to be a consequence of the failure of defense mechanisms and hence a hallmark of the disease. Oral bacteria detected in lower GIT have been linked to several pathological states like IBD, colorectal carcinoma [80], pancreatic ductal adenocarcinoma [81] or rheumatoid arthritis [82]. In an experiment, Klebsiella strains isolated from the saliva of human patients induced IBD in healthy germ-free mice [83]. Recently it has been shown at a large scale that despite oral-gut barriers, a substantial part of the oral microbes freely and frequently traverse the GIT and colonize different niches [84]. The transmissible bacteria included both pathogenic and commensal oral species [for example Prevotella strains or Fusobacterium nucleatum subspecies], however, the transmission scores were significantly increased for known opportunistic oral pathogens, causative agents of dental caries, and plaque-dwelling bacteria [84]. Endocarditis-associated species (Haemophilus, Aggregatibacter, Streptococcus) exhibited increased transmission scores as well. Taken together, this is an example how the microbiota originating from one niche may modify the composition of distant microbial communities.
The mechanism of migration of bacteria to out-of-GIT destination has not been fully elucidated yet but there is a growing body of evidence that alive bacteria could translocate through a leaky intestinal barrier and migrate via the circulation to the distal destinations [57]. Alternatively, the oral bacteria could reach the blood circulation system through minor injuries caused by tooth brushing o during dental treatment as formulated in the theory of focal infection reviewed recently by Olsen et al. [85]. Furthermore, the direct translocation of bacteria from one niche to another is not the only way how two or more microbial communities communicate and shape each other. At least two other mechanisms are well described, i.e. via the modulation of the host immune system and via bacterial fermentation products released into the circulation [86].
The dysbiosis precedes the clinical signs of the disease
In microbiota-related diseases, the dysbiosis often precedes the clinical signs of the disease [87, 88], and the shift in the microbiome composition could serve as a marker of the risk of the disease development. However, it remarkably challenges the definition of the “healthy microbiome”, because having no clinical signs of the disease does not automatically mean that the microbiome is not already dysbiotic.
An example of this phenomenon is the history of an effort to find a microbial signature of colorectal carcinoma. Numerous studies analyzed the gut microbiome in colorectal carcinoma patients with variable outcomes [89]. The most consistent result is the increased abundance of Fusobacterium nucleatum [90] both in the feces and in mucosa-associated with the tumor. Several other genera were reported to be either elevated [Peptostreptococcus, Streptococcus, Porphyromonas, Selenomonas, Enterococcus, Escheri-chia/Shigella, Klebsiella] or decreased [Roseburia, Lachnospiraceae] in colorectal carcinoma patients but the pattern was not uniform [91]. This controversy might be explained, at least partly, by the fact that the colorectal carcinoma-associated microbiome is being studied in a situation when the malignant conversion already occurred. For ethical reasons, it is difficult or even impossible to study the colon carcinogenesis “from the beginning” in humans. Therefore, we cannot be sure whether the observed alterations in the microbiome composition is the cause or the consequence of the cancer [92].
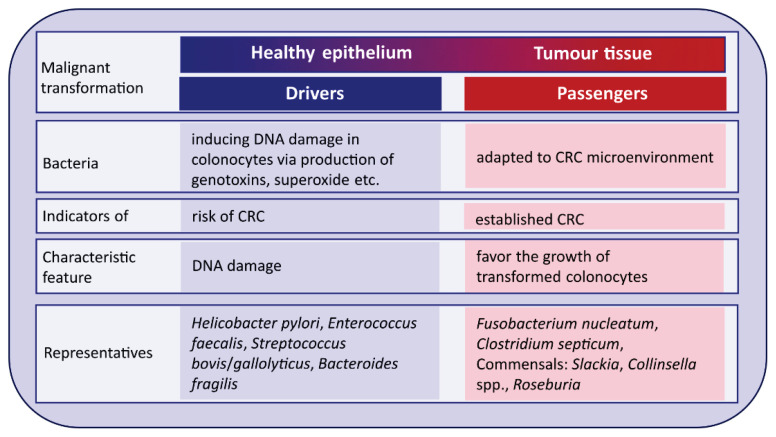
The driver-passenger model has been proposed by Tjalsma et al. [93] to explain steps leading to malignant conversion of colon epithelium and the role of bacteria in this process (Fig. 5). In this model, there are bacterial drivers and passengers, which contain bacteria with similar effects [94]. Several specific bacteria, the “drivers”, with pro-carcinogenic features initiate colorectal carcinoma development and start the process of malignant transformation of the healthy epithelium into tumor tissue. These key pathogens disappear as they failed to compete with opportunistic bacteria called “bacterial passengers” that are better adapted to the microenvironment of human colorectal carcinoma tumors [95]. Therefore, bacterial drivers can be considered as an indicator of a high risk of colorectal carcinoma, while the disappearance of bacterial drivers and the appearance of bacterial passengers may be indicators of the already established colorectal carcinoma [94]. So far, several “drivers” and “passengers” species have been proposed. Heliobacter pylori, Enterococcus faecalis, Streptococcus bovis/gallolyticus, and enterotoxigenic strains of Bacteroides fragilis are representatives of the “drivers” [96]. Bacterial “passengers” are bacteria well-adapted to the tumor microenvironment that in turn produce metabolites favoring the growth of transformed colonocytes. A characteristic feature of “passenger” bacteria is formation of biofilms what substantially increases their viability and provides them competitive advantage over non-aggregated microorganisms. The typical biofilm-forming “passenger” bacteria is Fusobac-terium nucleatum, which hampers the growth of butyrate-producing bacteria and thus reduces the release of butyrate, one of the main anticancer bacterial metabolites [92].
Fig. 5.
Drivers – passengers model. Adapted according to Tjalsma et al. [93].
The described example illustrate the motto of this paragraph “dysbiosis precedes the onset of the disease”. In the case of colorectal carcinoma, the first events promoting tumorigenesis occur in the restricted area of the gut and predominantly low-abundant mucosa-associated bacteria are involved. The dysbiosis is local and in the first stages, it is not projected into easily accessible fecal microbiota and the disease is still not overtly manifested. The microbiota associated with fully developed tumor may not be in causative relationship with the disease onset and merely reflects the altered state in the malignant tissue.
One size does not fit all
The vast majority of the microbes inhabiting various human body niches balance between commensalism [one partner benefits while the other is apparently unaffected] and mutualism [co-dependence among symbionts, in which both partners experience increased fitness] [97], some cause harm only under specific circumstances [opportunistic pathogens] and only few are currently considered to be strictly pathogenic. The actual relationship between the particular microorganism and the host depends on many conditions and what is beneficial in one setting may become detrimental in a different context. A growing body of information describing the multifaceted relationship among hosts and their microbial dwellers suggests that mutualism and pathogenicity are two sides of the same coin [22] and the actual interrelationship depends on the context.
Here we bring several examples that “one size does not fit all”. In a landmark study, Riquelme et al. showed that pancreatic adenocarcinoma tumors have a specific microbiome [103]. This microbiome is derived from the gut microbiota and more importantly, the tumor microbiome composition differs in patients with long- and short-term survival [97]. One of the key components of long-term survival tumor microbiota, Saccharopolyspora, was implicated in the inflammatory lung disease and was associated with cytokine overproduction [98]. The authors suggested that tumor microbiota associated with long-term survival contributes to the anti-tumor immune response by favoring recruitment and activation of CD8+ T cells, i.e. by inducing a pro-inflammatory immune response within the tumor microenvironment. Thus, in the context of pancreatic adenocarcinoma, the pro-inflammatory microbiota pattern, usually and justly considered unhealthy, brings a literally life-saving advantage to the host.
Most of the human gut bacteria possess the genetic equipment allowing for fermentation of substrates inaccessible to the host and thus increase the energy extracted from the food - but some strains are more efficient than others. In an elegant series of experiments on mono-colonized mice, Schwarzer et al. demonstrated that Lactobacillus plantarum promotes juvenile growth and mooreover, it buffered the adverse effects of chronic undernutrition [99]. Therefore, having these Lactobacillus strains in the gut microbiota may represent an advantage if the host faces the risk of malnutrition; however, it is a substantial disadvantage when the energy is in excess.
The gut microbiome is being adapted to the prevailing diet and lifestyle of the host. Few studies addressed the gut microbiome of still surviving communities of hunters and gatherers, among them Hadza people living in Tanzania [100]. The diet of the Hadza is very rich in diverse plant polysaccharides but low in amino acids. Compared to the urban communities living in Italy or USA, their microbiome is enriched in several bacterial genera including Prevotella. Prevotella species possess the enzymatic capacity to degrade carbohydrates and have a high capacity for branched-chain amino acid (BCAA) biosynthesis [101,102]. BCAA are essential amino acids that must be supplemented as food or from bacterial metabolism [103]. In the natural Hadza environment, Prevotella provides their hosts an advantage by increasing their capacity to process a vast array of refractory and resistant plant polysaccharides and supplementing BCAA missing in the diet.
At the same time, Prevotella may represent a health risk for the people living in urban areas. There is a long-lasting evidence that elevated circulating BCAA associate with insulin resistance, obesity, and diabetes [104] and may even predict cancer development [105]. The association between Prevotella-rich gut microbiome and insulin resistance was demonstrated. This particular example illustrates how diverse are interactions between the host – microbiome - environment. In one setting, the metabolic equipment (BCAA biosynthesis) may represent either an evolutionary advantage (in case of low availability of animal proteins) or a risk factor (in the situation of protein overnutrition). High fibrinolytic capability may be of utmost importance (when most of the calories are obtained from plant polysaccharides not easily accessible to humans) or negligible factor (when fiber in the diet is rare).
Other examples can be found in oral microbiome studies. When comparing healthy community with periodontitis patients, in almost every sufficiently big cohort there are few outliers of both types – clinically healthy individuals with a clearly dysbiotic microbiome and on the other hand severely affected patients with “healthy microbiomes” [88]. The authors hypothesize that some individuals possess an over-reactive immune system that triggers the proinflammatory reaction to otherwise symbiotic bacteria while subject with a less reactive immune system are more tolerant to pathogens. Several other examples of situations when people do not develop the same level of oral disease under the same circumstances are discussed in the review by Rosier et al. [54].
How to describe the microbiome
As mentioned above, the microbiome is a complex and dynamic structure and the choice of appropriate measures is a challenging task. We can ask about its taxonomic composition (“Who is present? How abundant is each component?”), about the functional potential (“What are the consortium members able to do?”), about their actual metabolic performance (“What are they doing just now?”) or how is the community stable or vulnerable.
The taxonomic composition could be addressed in principle by two approaches, 16S rRNA gene sequencing or shotgun metagenomic sequencing [WMS] each of them answering a somewhat different question. 16S rRNA gene sequencing provides, rapidly and for relatively low cost, information about taxonomic composition with limited precision and depth of identification. WMS informs us not only about the presence of individual taxa but also about the metabolic potential of the community, i.e. the presence of respective marker genes representing metabolic pathways, however, for the sake of higher costs and requirements for advanced computational skills [106]. An alternative approach is RNA sequencing which is similar to WMS in the principle, just instead of the microbial DNA, mRNA serves as a template. RNA sequencing identifies only genes that are actively transcribed at the time of sampling, i.e. it takes into consideration only the alive microorganisms and informs about their functional profile [106].
Most bacteria possess a wide metabolic repertoire and individual metabolic pathways could be easily switched on and off to maximize the energy yield from the available substrate[s]. Therefore, the same bacteria are capable to produce a very different spectrum of metabolites. The simple list of bacteria present in a sample or the metagenomic analysis including a list of encoded enzymes/metabolic pathways thus provide only partial information about the actual state of the studied community. In contrast, the metabolome has been proposed as a functional read-out of the human microbiome [107], reflective of microbiome–host interactions with an immediate impact on host health. Metabolomics identifies already biosynthesized metabolites/small molecules and therefore provides reliable information about the performance of the microbiota as a whole. On the other hand, we cannot assign particular metabolites to specific members of the consortium or to the host, and there are several other technological biases. There are two main approaches to metabolome analysis – targeted and untargeted. The targeted analysis focused on the preselected group of metabolites ensures the high reproducibility and accuracy of the outcome but the obtained information is limited to a narrow spectrum of compounds. The untargeted analysis aims to identify as many compounds as possible allowing for the elimination of selection bias. At the same time, this approach faces several limitations. First, the identification of hundreds of compounds is laborious, time-consuming, and sometimes impossible. Second, the selection of the sample processing and separation methods always limits the outputs only to part of the present metabolites. Third, the quantification of the obtained signals is complicated and usually, the quantity of a particular compound could be expressed only as a portion of the total, i.e. in percent, but not in absolute concentrations.
All the above-mentioned methodological approaches – metagenomics, metatranscriptomics, metabolomics – share one common feature, they produce a huge amount of data. The enormous technological development somewhat outruns our tools and ability to understand, visualize and interpret this reality and seriously complicates the integration of outcomes from different studies. The complexity of the microbiome systems impose enormous demands on the whole research pipeline what results in the reproducibility crisis [108]. Searching for the roots of this problem numerous studies were undertaken and unraveled that the chosen method significantly influence the outcome at virtually each step of the experimental procedure – from sample collection and DNA extraction [109], library preparation [110] to the bioinformatics pipeline [111] and data handling method [112]. In response to this challenge, guidelines for “wet lab procedures” (MBQC project, IHMS project) were established [110,113]. Standard guidelines tailored to microbiome study reporting called STORMS (“Strengthening The Organizing and Reporting of Microbiome Studies”) checklist were developed by a consortium of multidisciplinary specialists [114]. STORMS provides a tool to organize study planning and manuscript preparation, to improve the clarity of manuscripts, and to facilitate reviewers and readers in assessing these studies. Unfortunately, there is no general consensus on how to handle omics data on bioinformatics level so far and several approaches exist, all of them having their plus and cons [115]. The authors would like to stress that the selection of bioinformatics method and biostatistical approach always determines the outcome. At present, the only solution of this bottleneck is the openness in sharing the original sequencing data with sufficient metadata allowing for their re-analysis.
So far, we addressed only the cross-sectional description of the microbial community, i.e. “here and now”. Aiming to the description of a healthy microbiome, whatever it is, the more important issue is the assessment of sustainability measures of the microbial ecosystem, its stability, resistance, and resilience. Unfortunately, this field is still at the very beginning and the development of new methodological approaches is highly needed.
Examples of healthy microbiomes
All the above-mentioned facts make the postulation of the healthy microbiome of a specific human body site uncertain and complicated. Nevertheless, in few cases the scientists succeeded at least to describe the taxa as generally beneficial for their hosts, which thus could be considered a healthy microbiome of the niche.
Vaginal microbiome
The best example could be a relatively simple vaginal microbiome [116]. During the reproductive age, it is mainly dominated by Lactobacillus sp. which metabolites keep low pH protecting thus the genital tract and fetus from pathogenic microorganisms, for review see [117]. It can be affected by ethnicity [118], age, and hormonal state - negligibly by menstrual cycle [55] but remarkably during puberty and pregnancy [119]. In pregnancy, the species richness generally decreases [116] but the alpha diversity depends on the gestation week and could serve even as a predictive marker of the pre-term delivery risk [120]. The human vaginal microbiota is generally assigned to several vaginotypes or community state types (CSTs), first described by Ravel et al. [121], but following scientific papers in the field differ in the number of identified CSTs as well as in their characterization, which is always dependent on the clustering analysis of the entire evaluated sample set.
Nevertheless, we can conclude, that the vaginal microbiome of healthy adult women is predominantly composed of one or more Lactobacillus sp. and that some small percentage of women harbor a mixed population of non-Lactobacillus species based on Gardnerella vaginalis, Prevotella, Atopobium, Klebsiella and others. The Lactiobacillus sp.-based CSTs are considered beneficial [keeping low pH and producing metabolites protective against urogenital infections] while the mixed Gardnerella-based CST can indicate the risk of bacterial vaginosis. Among Lactobacillus-based CSTs, the predominance of L. crispatus in pregnancy is considered protective against the risk of preterm delivery, while the L. inners seems to indicate an increased risk of prematurity as well as the mixed Gardnerella based CST.
Oral microbiome
A much more complicated situation is in the oral cavity. After the gut, the oral cavity has the second largest and diverse microbiota [122]. It even gained its own database HOMD (Human oral microbiome database; https://www.homd.org/) harboring currently 774 oral bacterial species, 26 % of them being known only as uncultivated phylotypes [123]. Last, but not least, in the majority of scientific studies employing 16S rDNA-based taxonomy and clustering analysis comparing variable healthy and diseased groups, there are outliers, i.e. clearly diseased patients with the seemingly “healthy” microbiome and vice versa. For all these reasons, the estimation of the healthy oral microbiome is extremely tricky and it is clear, that “one size does not fit all”. Nevertheless, the current state of knowledge enables us to define at least some characteristics of beneficial oral microbiome of Caucasian individuals living in developed countries.
The healthy oral microbiome is generally based on variable species of Streptococcus, mainly S. mitis, S. oralis, S. gordonii, S. sanguinis or S. parasanquinis (S. mutans is associated with dental caries so it cannot be considered beneficial); further various Haemophillus species, Neisseria, Rothia, Gemella, Lautropia and probably also Veillonella [88], which, as an anaerobic microorganism, could be considered a transient taxon on the way to dysbiosis. Such oral microbiome often comprises also Fusobacterium nucleatum, which cannot be considered beneficial but its low percentage in oral cavity probably does not cause any harm (however, its presence in GIT is associated e.g. with increased risk of colon cancer) [81, 92].
However, when these more-less aerobic species are gradually replaced by F. nucleatum, Porphyromonas sp. like P. pasteri and P. catoniae, and Capnocytophaga sp., the microenvironment becomes more suitable for true periopathogenic taxa like red-complex bacteria Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, Fretibacterium sp. and Filifactor alocis. Such oral microbiome is considered dysbiotic and the respective individual is at a high risk of developing periodontal disease or is already symptomatic [88]. The interplay between the oral microbiome and immune system of the host is highly individual and the clear definition of the level of dysbiosis already critical for the development of the disease is not available. The tools enabling the evaluation of the dysbiosis based on the taxonomic composition of the oral microbiome, thus can be used to place the patient in question on a scale from health to the disease based on comparison with a database of already diagnosed individuals, however, it is only based on the statistic probability and there always would be some individuals misclassified [124].
Summary
An overwhelming amount of evidence proves that the human microbiome fully deserves to be considered an additional organ of the human body. Unfortunately, we still lack the appropriate measures allowing for the objective evaluation of whether the individual microbiome is healthy or not. Even the term “healthy” is misleading. It would be more appropriate to assess whether the microbiome composition and performance are (dis)advantageous for the host. The suitability of the particular microbiome composition for the host is always dynamic and depends on the situation of the host and the conditions of the environment; therefore, it is impossible to define one idealized community of specific microbes. The more promising approach may be to concentrate our effort on the definition of the essential [core] set of functions and metabolic modules that a healthy holobiont should possess – no matter if provided by its prokaryotic or eukaryotic part. Their absence could be predictive of the disease onset, especially in cases when the dysbiosis precedes the manifestation of the clinical symptoms. The therapeutic interventions should rather be focused on the replenishment of the attenuated/missing functions of the microbiome than on the simple provision of selected probiotic strains.
Furthermore, one of the key characteristics of a healthy microbiome is its resilience, i.e. the ability to maintain the necessary function in the changing environment even when it means the reorganization and changes in the composition of the community. The disturbations imposed on the human microbiome ecosystem are in most cases inevitable. Our efforts to reduce the resulting undesired shifts in the microbiome structure should preferentially address and strengthen the resilience rather than try to achieve some ideal composition.
Finally, to our opinion, the human microbiome must be envisioned as a complex system tightly interconnected with other macro- and micro-ecosystems in our environment. Our, i.e. human, microbiome cannot stay healthy in an otherwise unhealthy environment, and therefore, it is essential to pay similar attention to all components of the planetary ecosystem.
Acknowledgements
This study was supported by the Ministry of Health of the Czech Republic, grant NV-18-01-00040, by the project National Institute of virology and bacteriology (Programme EXCELES, ID Project No. LX22NPO5103) - Funded by the European Union - Next Generation EU and by the project National Institute for Research of Metabolic and Cardiovascular Diseases (Programme EXCELES, Project No. LX22NPO5104) - Funded by the European Union - Next Generation EU.
Footnotes
Conflict of Interest
There is no conflict of interest.
References
- 1.Gordon J, Knowlton N, Relman DA, Rohwer F, Youle M. Superorganisms and holobionts. Microbe. 2013;8:152–3. doi: 10.1128/microbe.8.152.1. [DOI] [Google Scholar]
- 2.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert SF. A holobiont birth narrative: the epigenetic transmission of the human microbiome. Front Genet. 2014;5:282. doi: 10.3389/fgene.2014.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levin Polasky S, Simon A. Fragile Dominion: Complexity and the Commons. Reading MA: Perseus Books; 1999. p. 254. [Google Scholar]; American Journal of Agricultural Economics. 2001;83(1):246–7. doi: 10.1111/1467-8276.t01-1-00151. [DOI] [Google Scholar]
- 5.Carl F, Steve C, Brian W, Marten S, Thomas E, Lance G, et al. Regime Shifts, Resilience, and Biodiversity in Ecosystem Management. Annual Review of Ecology, Evolution, and Systematics. 2004;35(1):557–81. doi: 10.1146/annurev.ecolsys.35.021103.105711. [DOI] [Google Scholar]
- 6.Relman DA. The human microbiome: ecosystem resilience and health. Nutr Rev. 2012;70(Suppl 1):S2–9. doi: 10.1111/j.1753-4887.2012.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pimm SL. Ecological Issues in the Conservation of Species and Communities. University of Chicago Press; 1991. The Balance of Nature? [Google Scholar]
- 9.Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15(10):630–8. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 10.Holling CS. Resilience and Stability of Ecological Systems. Annual Review of Ecology and Systematics. 1973;4(1):1–23. doi: 10.1146/annurev.es.04.110173.000245. [DOI] [Google Scholar]
- 11.Moustafa A, Li W, Anderson EL, Wong EHM, Dulai PS, Sandborn WJ, et al. Genetic risk, dysbiosis, and treatment stratification using host genome and gut microbiome in inflammatory bowel disease. Clin Transl Gastroenterol. 2018;9(1):e132. doi: 10.1038/ctg.2017.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407–15. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 13.McNaughton SJ. Diversity and Stability of Ecological Communities: A Comment on the Role of Empiricism in Ecology. The American Naturalist. 1977;111(979):515–25. doi: 10.1086/283181. [DOI] [Google Scholar]
- 14.Naeem S, Li S. Biodiversity enhances ecosystem reliability. Nature. 1997;390(6659):507–9. doi: 10.1038/37348. [DOI] [Google Scholar]
- 15.Yachi S, Loreau M. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc Natl Acad Sci U S A. 1999;96(4):1463–8. doi: 10.1073/pnas.96.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber M, Knottnerus JA, Green L, van der Horst H, Jadad AR, Kromhout D, et al. How should we define health? BMJ. 2011;343:d4163. doi: 10.1136/bmj.d4163. [DOI] [PubMed] [Google Scholar]
- 17.Oleribe OO, Ukwedeh O, Burstow NJ, Gomaa AI, Sonderup MW, Cook N, et al. Health: redefined. Pan Afr Med J. 2018;30:292. doi: 10.11604/pamj.2018.30.292.15436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stokes J, 3rd, Noren J, Shindell S. Definition of terms and concepts applicable to clinical preventive medicine. J Community Health. 1982;8(1):33–41. doi: 10.1007/BF01324395. [DOI] [PubMed] [Google Scholar]
- 19.Dietert RR. Microbiome First Medicine in Health and Safety. Biomedicines. 2021;9(9) doi: 10.3390/biomedicines9091099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15(7):R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449(7164):811–8. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen OFA, van de Burgwal LHM. On the Verge of a Catastrophic Collapse? The Need for a Multi-Ecosystem Approach to Microbiome Studies. Front Microbiol. 2021;12:784797. doi: 10.3389/fmicb.2021.784797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107(33):14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banerjee S, Schlaeppi K, van der Heijden MGA. Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol. 2018;16(9):567–76. doi: 10.1038/s41579-018-0024-1. [DOI] [PubMed] [Google Scholar]
- 26.Bach JF. Revisiting the Hygiene Hypothesis in the Context of Autoimmunity. Front Immunol. 2020;11:615192. doi: 10.3389/fimmu.2020.615192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178–84. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 28.Voorhies AAL, HA The Challenge of Maintaining a Healthy Microbiome during Long-Duration Space Missions. Front Astron Space Sci. 2016;3(23):1–7. doi: 10.3389/fspas.2016.00023. [DOI] [Google Scholar]
- 29.Bach JF. The hygiene hypothesis in autoimmunity: the role of pathogens and commensals. Nat Rev Immunol. 2018;18(2):105–20. doi: 10.1038/nri.2017.111. [DOI] [PubMed] [Google Scholar]
- 30.Rook GA. Review series on helminths, immune modulation and the hygiene hypothesis: the broader implications of the hygiene hypothesis. Immunology. 2009;126(1):3–11. doi: 10.1111/j.1365-2567.2008.03007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Hertzen L, Hanski I, Haahtela T. Natural immunity. Biodiversity loss and inflammatory diseases are two global megatrends that might be related. EMBO Rep. 2011;12(11):1089–93. doi: 10.1038/embor.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kostovcikova K, Coufal S, Galanova N, Fajstova A, Hudcovic T, Kostovcik M, et al. Diet rich in animal protein promotes pro-inflammatory macrophage response and exacerbates colitis in mice. Front Immunol. 2019;10:919. doi: 10.3389/fimmu.2019.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Dong Y, Sun G, Hasan AA, Tian M, Zeng S, et al. Paternal programming of liver function and lipid profile induced by a paternal pre-conceptional unhealthy diet: potential association with altered gut microbiome composition. Kidney Blood Press Res. 2019;44(1):133–48. doi: 10.1159/000497487. [DOI] [PubMed] [Google Scholar]
- 34.Sun CY, Zheng ZL, Chen CW, Lu BW, Liu D. Targeting gut microbiota with natural polysaccharides: effective interventions against high-fat diet-induced metabolic diseases. Front Microbiol. 2022;13:859206. doi: 10.3389/fmicb.2022.859206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panelli S, Epis S, Cococcioni L, Perini M, Paroni M, Bandi C, et al. Inflammatory bowel diseases, the hygiene hypothesis and the other side of the microbiota: Parasites and fungi. Pharmacol Res. 2020;159:104962. doi: 10.1016/j.phrs.2020.104962. [DOI] [PubMed] [Google Scholar]
- 36.Fyhrquist N. The human microbiota and its relationship with allergies. Gastroenterol Clin North Am. 2019;48(3):377–87. doi: 10.1016/j.gtc.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 37.Olunoiki E, Rehner J, Bischoff M, Koshel E, Vogt T, Reichrath J, et al. Characteristics of the Skin Microbiome in Selected Dermatological Conditions: A Narrative Review. Life (Basel) 2022;12(9) doi: 10.3390/life12091420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tramper-Stranders G, Ambrozej D, Arcolaci A, Atanaskovic-Markovic M, Boccabella C, Bonini M, et al. Dangerous liaisons: Bacteria, antimicrobial therapies, and allergic diseases. Allergy. 2021;76(11):3276–91. doi: 10.1111/all.15046. [DOI] [PubMed] [Google Scholar]
- 39.Matenchuk BA, Mandhane PJ, Kozyrskyj AL. Sleep, circadian rhythm, and gut microbiota. Sleep Med Rev. 2020;53:101340. doi: 10.1016/j.smrv.2020.101340. [DOI] [PubMed] [Google Scholar]
- 40.Frazier K, Chang EB. Intersection of the Gut Microbiome and Circadian Rhythms in Metabolism. Trends Endocrinol Metab. 2020;31(1):25–36. doi: 10.1016/j.tem.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pickard JM, Zeng MY, Caruso R, Nunez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279(1):70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Datar A, Nicosia N. Assessing Social Contagion in Body Mass Index, Overweight, and Obesity Using a Natural Experiment. JAMA Pediatr. 2018;172(3):239–46. doi: 10.1001/jamapediatrics.2017.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christakis NA, Fowler JH. The spread of obesity in a large social network over 32 years. N Engl J Med. 2007;357(4):370–9. doi: 10.1056/NEJMsa066082. [DOI] [PubMed] [Google Scholar]
- 45.Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67(9):1716–25. doi: 10.1136/gutjnl-2018-316723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gregory JC, Buffa JA, Org E, Wang Z, Levison BS, Zhu W, et al. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem. 2015;290(9):5647–60. doi: 10.1074/jbc.M114.618249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145(5):745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thaiss CA, Itav S, Rothschild D, Meijer MT, Levy M, Moresi C, et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature. 2016;540(7634):544–51. doi: 10.1038/nature20796. [DOI] [PubMed] [Google Scholar]
- 49.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walter J, Armet AM, Finlay BB, Shanahan F. Establishing or Exaggerating Causality for the Gut Microbiome: Lessons from Human Microbiota-Associated Rodents. Cell. 2020;180(2):221–32. doi: 10.1016/j.cell.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 51.Bajaj JS, Ng SC, Schnabl B. Promises of microbiome-based therapies. J Hepatol. 2022;76(6):1379–91. doi: 10.1016/j.jhep.2021.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marotz CA, Zarrinpar A. Treating Obesity and Metabolic Syndrome with Fecal Microbiota Transplantation. Yale J Biol Med. 2016;89(3):383–8. [PMC free article] [PubMed] [Google Scholar]
- 53.Alang N, Kelly CR. Weight gain after fecal microbiota transplantation. Open Forum Infect Dis. 2015;2(1):ofv004. doi: 10.1093/ofid/ofv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosier BT, Marsh PD, Mira A. Resilience of the Oral Microbiota in Health: Mechanisms That Prevent Dysbiosis. J Dent Res. 2018;97(4):371–80. doi: 10.1177/0022034517742139. [DOI] [PubMed] [Google Scholar]
- 55.Chaban B, Links MG, Jayaprakash TP, Wagner EC, Bourque DK, Lohn Z, et al. Characterization of the vaginal microbiota of healthy Canadian women through the menstrual cycle. Microbiome. 2014;2:23. doi: 10.1186/2049-2618-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oh J, Byrd AL, Deming C, Conlan S, Program NCS, Kong HH, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514(7520):59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagpal R, Yadav H. Bacterial Translocation from the Gut to the Distant Organs: An Overview. Ann Nutr Metab. 2017;71(Suppl 1):11–6. doi: 10.1159/000479918. [DOI] [PubMed] [Google Scholar]
- 58.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Toole PW, Jeffery IB. Gut microbiota and aging. Science. 2015;350(6265):1214–5. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- 60.Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16:90. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salazar N, Gonzalez S, Nogacka AM, Rios-Covian D, Arboleya S, Gueimonde M, et al. Microbiome: Effects of Ageing and Diet. Curr Issues Mol Biol. 2020;36:33–62. doi: 10.21775/cimb.036.033. [DOI] [PubMed] [Google Scholar]
- 62.Nagpal R, Mainali R, Ahmadi S, Wang S, Singh R, Kavanagh K, et al. Gut microbiome and aging: Physiological and mechanistic insights. Nutr Healthy Aging. 2018;4(4):267–85. doi: 10.3233/NHA-170030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rios-Covian D, Gonzalez S, Nogacka AM, Arboleya S, Salazar N, Gueimonde M, et al. An Overview on Fecal Branched Short-Chain Fatty Acids Along Human Life and as Related With Body Mass Index: Associated Dietary and Anthropometric Factors. Front Microbiol. 2020;11:973. doi: 10.3389/fmicb.2020.00973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trivedi B. Microbiome: The surface brigade. Nature. 2012;492(7429):S60–1. doi: 10.1038/492S60a. [DOI] [PubMed] [Google Scholar]
- 65.Bay L, Barnes CJ, Fritz BG, Thorsen J, Restrup MEM, Rasmussen L, et al. Universal Dermal Microbiome in Human Skin. mBio. 2020;11(1) doi: 10.1128/mBio.02945-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Humphrey SP, Williamson RT. A review of saliva: normal composition, flow, and function. J Prosthet Dent. 2001;85(2):162–9. doi: 10.1067/mpr.2001.113778. [DOI] [PubMed] [Google Scholar]
- 67.Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016;14(8):e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berthelot JM, Bandiaky ON, Le Goff B, Amador G, Chaux AG, Soueidan A, et al. Another Look at the Contribution of Oral Microbiota to the Pathogenesis of Rheumatoid Arthritis: A Narrative Review. Microorganisms. 2021;10(1) doi: 10.3390/microorganisms10010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khan I, Khan I, Jianye Z, Xiaohua Z, Khan M, Hilal MG, et al. Exploring blood microbial communities and their influence on human cardiovascular disease. J Clin Lab Anal. 2022;36(4):e24354. doi: 10.1002/jcla.24354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martinez M, Postolache TT, Garcia-Bueno B, Leza JC, Figuero E, Lowry CA, et al. The role of the oral microbiota related to periodontal diseases in anxiety, mood and trauma- and stress-related disorders. Front Psychiatry. 2021;12:814177. doi: 10.3389/fpsyt.2021.814177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Newman KL, Kamada N. Pathogenic associations between oral and gastrointestinal diseases. Trends Mol Med. 2022 doi: 10.1016/j.molmed.2022.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sumida K, Han Z, Chiu CY, Mims TS, Bajwa A, Demmer RT, et al. Circulating Microbiota in Cardiometabolic Disease. Front Cell Infect Microbiol. 2022;12:892232. doi: 10.3389/fcimb.2022.892232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15(1):30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salem I, Ramser A, Isham N, Ghannoum MA. The Gut Microbiome as a Major Regulator of the Gut-Skin Axis. Front Microbiol. 2018;9:1459. doi: 10.3389/fmicb.2018.01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sanchez-Pellicer P, Navarro-Moratalla L, Nunez-Delegido E, Ruzafa-Costas B, Aguera-Santos J, Navarro-Lopez V. Acne, Microbiome, and Probiotics: The Gut-Skin Axis. Microorganisms. 2022;10(7) doi: 10.3390/microorganisms10071303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chhibber-Goel J, Singhal V, Bhowmik D, Vivek R, Parakh N, Bhargava B, et al. Linkages between oral commensal bacteria and atherosclerotic plaques in coronary artery disease patients. NPJ Biofilms Microbiomes. 2016;2:7. doi: 10.1038/s41522-016-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williams JE, Carrothers JM, Lackey KA, Beatty NF, Brooker SL, Peterson HK, et al. Strong Multivariate Relations Exist Among Milk, Oral, and Fecal Microbiomes in Mother-Infant Dyads During the First Six Months Postpartum. J Nutr. 2019;149(6):902–14. doi: 10.1093/jn/nxy299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Poole S, Singhrao SK, Kesavalu L, Curtis MA, Crean S. Determining the presence of periodontopathic virulence factors in short-term postmortem Alzheimer’s disease brain tissue. J Alzheimers Dis. 2013;36(4):665–77. doi: 10.3233/JAD-121918. [DOI] [PubMed] [Google Scholar]
- 79.Xu B, Han YW. Oral bacteria, oral health, and adverse pregnancy outcomes. Periodontol 2000. 2022;89(1):181–9. doi: 10.1111/prd.12436. [DOI] [PubMed] [Google Scholar]
- 80.Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6:8727. doi: 10.1038/ncomms9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6(9):7209–20. doi: 10.18632/oncotarget.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21(8):895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 83.Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science. 2017;358(6361):359–65. doi: 10.1126/science.aan4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schmidt TS, Hayward MR, Coelho LP, Li SS, Costea PI, Voigt AY, et al. Extensive transmission of microbes along the gastrointestinal tract. Elife. 2019:8. doi: 10.7554/eLife.42693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olsen I, van Winkelhoff AJ. Acute focal infections of dental origin. Periodontol 2000. 2014;65(1):178–89. doi: 10.1111/prd.12018. [DOI] [PubMed] [Google Scholar]
- 86.Park SY, Hwang BO, Lim M, Ok SH, Lee SK, Chun KS, et al. Oral-Gut Microbiome Axis in Gastrointestinal Disease and Cancer. Cancers (Basel) 2021;13(9) doi: 10.3390/cancers13092124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One. 2012;7(6):e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lenartova M, Tesinska B, Janatova T, Hrebicek O, Mysak J, Janata J, et al. The Oral Microbiome in Periodontal Health. Front Cell Infect Microbiol. 2021;11:629723. doi: 10.3389/fcimb.2021.629723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jahani-Sherafat S, Alebouyeh M, Moghim S, Ahmadi Amoli H, Ghasemian-Safaei H. Role of gut microbiota in the pathogenesis of colorectal cancer; a review article. Gastroenterol Hepatol Bed Bench. 2018;11(2):101–9. [PMC free article] [PubMed] [Google Scholar]
- 90.Lawrence GW, Begley M, Cotter PD, Guinane CM. Potential Use of Biotherapeutic Bacteria to Target Colorectal Cancer-Associated Taxa. Int J Mol Sci. 2020;21(3) doi: 10.3390/ijms21030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6(2):320–9. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ranjbar M, Salehi R, Haghjooy Javanmard S, Rafiee L, Faraji H, Jafarpor S, et al. The dysbiosis signature of Fusobacterium nucleatum in colorectal cancer-cause or consequences? A systematic review. Cancer Cell Int. 2021;21(1):194. doi: 10.1186/s12935-021-01886-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10(8):575–82. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 94.Xing J, Fang Y, Zhang W, Zhang H, Tang D, Wang D. Bacterial driver-passenger model in biofilms: a new mechanism in the development of colorectal cancer. Clin Transl Oncol. 2022;24(5):784–95. doi: 10.1007/s12094-021-02738-y. [DOI] [PubMed] [Google Scholar]
- 95.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Avril M, DePaolo RW. “Driver-passenger” bacteria and their metabolites in the pathogenesis of colorectal cancer. Gut Microbes. 2021;13(1):1941710. doi: 10.1080/19490976.2021.1941710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell. 2019;178(4):795–806 e12. doi: 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim YI, Park JE, Brand DD, Fitzpatrick EA, Yi AK. Protein kinase D1 is essential for the proinflammatory response induced by hypersensitivity pneumonitis-causing thermophilic actinomycetes Saccharopolyspora rectivirgula. J Immunol. 2010;184(6):3145–56. doi: 10.4049/jimmunol.0903718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schwarzer M, Makki K, Storelli G, Machuca-Gayet I, Srutkova D, Hermanova P, et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science. 2016;351(6275):854–7. doi: 10.1126/science.aad8588. [DOI] [PubMed] [Google Scholar]
- 100.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dahl WJ, Rivero Mendoza D, Lambert JM. Diet, nutrients and the microbiome. Prog Mol Biol Transl Sci. 2020;171:237–63. doi: 10.1016/bs.pmbts.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 102.Yue SJ, Liu J, Wang AT, Meng XT, Yang ZR, Peng C, et al. Berberine alleviates insulin resistance by reducing peripheral branched-chain amino acids. Am J Physiol Endocrinol Metab. 2019;316(1):E73–E85. doi: 10.1152/ajpendo.00256.2018. [DOI] [PubMed] [Google Scholar]
- 103.Gojda J, Cahova M. Gut Microbiota as the Link between Elevated BCAA Serum Levels and Insulin Resistance. Biomolecules. 2021;11(10) doi: 10.3390/biom11101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–26. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Katagiri R, Goto A, Nakagawa T, Nishiumi S, Kobayashi T, Hidaka A, et al. Increased Levels of Branched-Chain Amino Acid Associated With Increased Risk of Pancreatic Cancer in a Prospective Case-Control Study of a Large Cohort. Gastroenterology. 2018;155(5):1474–82 e1. doi: 10.1053/j.gastro.2018.07.033. [DOI] [PubMed] [Google Scholar]
- 106.Wensel CR, Pluznick JL, Salzberg SL, Sears CL. Next-generation sequencing: insights to advance clinical investigations of the microbiome. J Clin Invest. 2022;132(7) doi: 10.1172/JCI154944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zierer J, Jackson MA, Kastenmuller G, Mangino M, Long T, Telenti A, et al. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet. 2018;50(6):790–5. doi: 10.1038/s41588-018-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schloss PD. Identifying and Overcoming Threats to Reproducibility, Replicability, Robustness, and Generalizability in Microbiome Research. mBio. 2018;9(3) doi: 10.1128/mBio.00525-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Greathouse KL, Sinha R, Vogtmann E. DNA extraction for human microbiome studies: the issue of standardization. Genome Biol. 2019;20(1):212. doi: 10.1186/s13059-019-1843-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Costea PI, Zeller G, Sunagawa S, Pelletier E, Alberti A, Levenez F, et al. Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol. 2017;35(11):1069–76. doi: 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- 111.O’Sullivan DM, Doyle RM, Temisak S, Redshaw N, Whale AS, Logan G, et al. An inter-laboratory study to investigate the impact of the bioinformatics component on microbiome analysis using mock communities. Sci Rep. 2021;11(1):10590. doi: 10.1038/s41598-021-89881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Clausen DS, Willis AD. Evaluating replicability in microbiome data. Biostatistics. 2022;23(4):1099–114. doi: 10.1093/biostatistics/kxab048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sinha R, Abu-Ali G, Vogtmann E, Fodor AA, Ren B, Amir A, et al. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat Biotechnol. 2017;35(11):1077–86. doi: 10.1038/nbt.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mirzayi C, Renson A, Massive A, Zohra F, et al. Quality Control S, Genomic Standards C. Reporting guidelines for human microbiome research: the STORMS checklist. Nat Med. 2021;27(11):1885–92. doi: 10.1038/s41591-021-01552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nearing JT, Douglas GM, Hayes MG, MacDonald J, Desai DK, Allward N, et al. Microbiome differential abundance methods produce different results across 38 datasets. Nat Commun. 2022;13(1):342. doi: 10.1038/s41467-022-28034-z. https://doi.org/10.1038/s41467-022-28401-w, https://doi.org/10.1038/s41467-022-28401-whttps://doi.org/10.1038/s41467-022-28034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mancabelli L, Tarracchini C, Milani C, Lugli GA, Fontana F, Turroni F, et al. Vaginotypes of the human vaginal microbiome. Environ Microbiol. 2021;23(3):1780–92. doi: 10.1111/1462-2920.15441. [DOI] [PubMed] [Google Scholar]
- 117.Verstraelen H, Vieira-Baptista P, De Seta F, Ventolini G, Lonnee-Hoffmann R, Lev-Sagie A. The Vaginal Microbiome: I. Research Development, Lexicon, Defining “Normal” and the Dynamics Throughout Women’s Lives. J Low Genit Tract Dis. 2022;26(1):73–8. doi: 10.1097/LGT.0000000000000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dominguez-Bello MG. Gestational shaping of the maternal vaginal microbiome. Nat Med. 2019;25(6):882–3. doi: 10.1038/s41591-019-0483-6. [DOI] [PubMed] [Google Scholar]
- 119.Auriemma RS, Scairati R, Del Vecchio G, Liccardi A, Verde N, Pirchio R, et al. The Vaginal Microbiome: A Long Urogenital Colonization Throughout Woman Life. Front Cell Infect Microbiol. 2021;11:686167. doi: 10.3389/fcimb.2021.686167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Haque MM, Merchant M, Kumar PN, Dutta A, Mande SS. First-trimester vaginal microbiome diversity: A potential indicator of preterm delivery risk. Sci Rep. 2017;7(1):16145. doi: 10.1038/s41598-017-16352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4680–7. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deo PN, Deshmukh R. Oral microbiome: Unveiling the fundamentals. J Oral Maxillofac Pathol. 2019;23(1):122–8. doi: 10.4103/jomfp.JOMFP_77_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP. New Insights into Human Nostril Microbiome from the Expanded Human Oral Microbiome Database (eHOMD): a Resource for the Microbiome of the Human Aerodigestive Tract. mSystems. 2018;3(6) doi: 10.1128/mSystems.00187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Najmanova L, Sabova L, Lenartova M, Janatova T, Mysak J, Vetrovsky T, et al. R/G Value-A Numeric Index of Individual Periodontal Health and Oral Microbiome Dynamics. Front Cell Infect Microbiol. 2021;11:602643. doi: 10.3389/fcimb.2021.602643. [DOI] [PMC free article] [PubMed] [Google Scholar]