Abstract
This study was done to elucidate the signal transduction pathway of interleukin-8 (IL-8) induction by gram-positive bacteria. Bacteria (micrococci) and peptidoglycan (PGN) induced transcription of IL-8 in HEK293 cells expressing Toll-like receptor 2 (TLR2) and CD14 but not in those expressing TLR1 or TLR4. A mutation within the NF-κB site in the IL-8 promoter abrogated transcriptional induction of IL-8 by the two stimulants. Dominant negative myeloid differentiation protein (MyD88), IL-1 receptor-associated kinase (IRAK), NFκB-inducing kinase (NIK), and IκB kinase (IKK) mutant forms completely inhibited micrococcus- and PGN-induced activation of NF-κB and expression of the gene for IL-8. Induction of NF-κB was partially inhibited by dominant negative tumor necrosis factor receptor-associated kinase 6 (TRAF6) but not TRAF2, whereas induction of IL-8 gene was partially inhibited by both TRAF6 and TRAF2. These data indicate that micrococci and PGN induce TLR2-dependent activation of the gene for IL-8 and that this activation requires MyD88, IRAK, NIK, IKK, and NF-κB and may also utilize TRAF6 and, to a lesser extent, TRAF2.
Innate immunity is an early line of host defense against pathogens. Both gram-positive and gram-negative bacteria and their cell wall components activate the innate immune system of the host and induce secretion of proinflammatory molecules, mainly chemokines and cytokines (1, 5–7, 21, 32). These inflammatory molecules are the main mediators of the pathological effects induced by bacteria, including inflammation, fever, hypotension, leukocytosis, decreased appetite, and arthritis (1, 5–7, 21).
We have recently discovered that chemokines are the main proinflammatory mediators induced in monocytes by bacteria and peptidoglycan (PGN) and lipopolysaccharide (LPS), the main cell wall components of gram-positive and gram-negative bacteria, respectively (32). The gene for the chemokine interleukin-8 (IL-8) is the gene most highly induced by all bacterial stimulants out of 600 genes studied (32). However, the mechanism of this induction, i.e., the receptors, signal transduction pathways, and transcription factors involved in the transcriptional activation of the gene for IL-8, are unknown.
Gram-positive bacteria and PGN activate cells through the pattern recognition receptors CD14 and Toll-like receptor 2 (TLR2), which results in the activation of the transcription factor NF-κB (8, 11, 27, 29–31, 36). NF-κB is a ubiquitous transcription factor that regulates the transcription of various genes involved in immune responses. LPS from gram-negative bacteria also induces activation of NF-κB through CD14 and TLR (16, 34), and this activation requires the signal transduction molecules myeloid differentiation protein (MyD88), IL-1 receptor-associated kinase (IRAK), tumor necrosis factor (TNF) receptor-associated kinase 6 (TRAF6), NF-κB-inducing kinase (NIK), and IκB kinase (IKK) (16, 30, 34, 37). LPS stimulation of this pathway results in the activation of IKK (10), which then phosphorylates IκB, resulting in its degradation and the subsequent release and translocation of NF-κB to the nucleus, where NF-κB activates various genes. However, the signal transduction pathway that is activated by gram-positive bacteria and PGN and results in the activation of NF-κB is not known and furthermore, the role of NF-κB in the induction of the gene for IL-8 (and the genes for other chemokines and cytokines) is also unknown.
Therefore, the objectives of this study were to determine whether gram-positive bacteria and their PGN component (i) induce TLR2-dependent transcription of IL-8, (ii) activate the TLR2→MyD88→IRAK→TRAF6→NIK→IKK→NF-κB signal transduction pathway, and (iii) activate the gene for IL-8 through this TLR2-mediated pathway.
MATERIALS AND METHODS
Materials.
Soluble PGN (sPGN), a polymeric un-cross-linked PGN (average Mr = 125,000) released from Staphylococcus aureus grown in the presence of penicillin, was purified by vancomycin affinity chromatography and analyzed as described before (25). Micrococcus luteus ATCC 4698 (obtained from Sigma, St. Louis, Mo.) was used as a prototypic gram-positive nonpathogenic (i.e., easily eliminated by the host innate immune system) bacterium with unmodified and unsubstituted PGN readily accessible on its surface. sPGN and micrococci contained <24 and <500 pg of endotoxin/mg, respectively, as determined by the Limulus lysate assay (25). Recombinant mouse TNF-α (specific activity, 4 × 107 U/ml in the L929 cytotoxicity assay; containing ≤1 endotoxin unit/mg; obtained from Genzyme, Boston, Mass.) was used as a control nonbacterial stimulus that activates NF-κB through a pathway that is initially distinct from the TLR- and IL-1R-induced pathways (26).
Cell culture.
The human embryonic kidney cell line HEK293 (American Type Culture Collection, Manassas, Va.) was cultured in Dulbecco's modified Eagle's medium with 10% defined fetal calf serum (HyClone, Logan, Utah; endotoxin content, <6 pg/ml). The stable transfectants expressing different TLRs with a FLAG epitope at the 5′ end or the control vector, 293/TLR1-5′, 293/TLR4-5′, 293/TLR2-5′, and 293/cv, were generated and cultured as described before (16, 27).
Electrophoretic mobility shift assays.
Cells were cultured at 0.35 × 106 to 0.4 × 106/ml in 24-well plates (1.0 ml/well) for 16 to 20 h and stimulated as indicated in the figure legends, and nuclear extracts were prepared as described before (11). Nuclear proteins (5 μg) were incubated with 32P-labeled oligonucleotide containing a consensus NF-κB binding site for 30 min at 22°C as described before (11). All samples were then separated on 6% nondenaturing polyacrylamide gels, and the DNA-protein complexes were visualized by autoradiography.
RNA isolation and reverse transcription-PCR.
293/TLR2-5′ cells were cultured at 0.75 × 106 to 1.0 × 106/ml in six-well plates (2.0 ml/well) for 16 to 20 h and stimulated as indicated in the figure legends, and total RNA was isolated using the RNeasy Purification Kit (Qiagen, Valencia, Calif.). One microgram of total RNA was used for the synthesis of cDNA, and the cDNA was then amplified using the Access kit (Promega, Madison, Wis.). The primers used for the amplification of IL-8 cDNA were (14) 5′ GCAGCTCTGTGTGAAGGTGCAGTTT 3′ (sense) and 5′ CTCAGCCCTCTTCAAAAACTTCTCC (antisense). The same antisense primer was used for the synthesis of IL-8 cDNA from total RNA. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as a control, and GAPDH cDNA was amplified with the primers 5′ ACCACAGTCCATGCCATCAC 3′ (sense) and 5′ TCCACCACCCTGTTGCTGTA 3′ (antisense). The same antisense primer was used for the synthesis of GAPDH cDNA from total RNA. Synthesis of cDNA was done at 48°C for 45 min, and the amplification was performed with an initial denaturation step of 94°C for 3 min, followed by 45 cycles of 55°C for 20 s, 72°C for 30 s, and 94°C for 20 s, and a final polymerizing step of 72°C for 7 min. The amplified products were separated on a 3% Nu-Sieve agarose gel, stained with ethidium bromide, and quantified using Kodak Digital Science Image Station 440CF and Image Analysis Software 3.0.
Transfection and chloramphenicol acetyltransferase (CAT) and luciferase assays.
HEK293 cells were cultured at 0.35 × 106 to 0.4 × 106/ml in 48-well plates (0.25 ml/well) for 16 to 20 h and transfected with Lipofectamine and DNA (the amount of DNA used was optimized for different plasmids and is indicated in the figure legends). Duplicate or triplicate wells were set up for each group. The reporter plasmids used were ELAM-1 luciferase (27) and IL8-CAT (17). The plasmids expressing CD14 (5), TLR1 (27), TLR2 (27), and TLR4 (27) have been described previously. Expression of surface molecules was monitored by Western blotting of cell lysates with antitag antibodies as previously described (8). The following plasmids expressing dominant negative mutant proteins were used for transfections: MyD88[152-296] (20), ΔIRAK1 (20), IRAK2[97-590] (20), TRAF6[289-522] (4), TRAF2[87-501] (26), NIK[KK429,430AA] (33), IKKα[S176A] (24), IKKβ[S177A] (24), and IκBΔN (3). Cells were allowed to recover for 12 to 24 h and then were left unstimulated or were stimulated as described in the figure legends. Lysates were prepared and were assayed for luciferase activity using the Luciferase Reporter kit (Promega) or for CAT activity as previously described (12).
RESULTS
Micrococci and sPGN induce TLR2-dependent transcription of the gene for IL-8 in HEK293 cells, and this induction requires NF-κB.
Because the gene for IL-8 is the most highly induced gene for proinflammatory molecules in human monocytes activated by bacteria and bacterial products (32) and because PGN and gram-positive bacteria activate cells through TLR2, we wanted to test the hypothesis that the transcriptional activation of the gene for IL-8 is mediated through TLR2. We tested whether micrococci and sPGN induce TLR2-dependent transcription of the gene for IL-8 by two different assays: (i) reverse transcription-PCR of IL-8 mRNA in HEK293 cells stably transfected with TLR2 and (ii) transactivation of an IL-8 promoter–CAT construct in HEK293 cells transiently transfected with pIL8(wt)CAT and with plasmids expressing TLR2 and CD14.
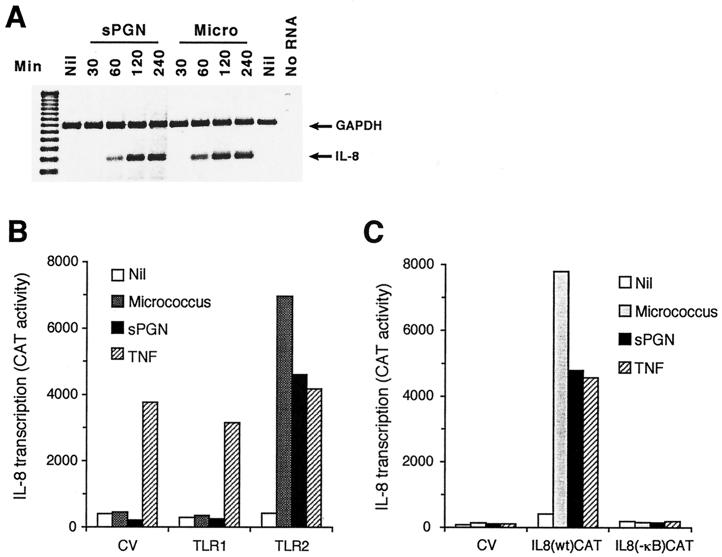
IL-8 mRNA was induced within 60 min of activation and continued to increase for up to 4 h after activation by micrococci and sPGN (Fig. 1A). IL-8 cDNA and the control GAPDH cDNA were synthesized and amplified as 243- and 451-bp fragments, respectively. This induction was specific for IL-8 because the amount of GAPDH mRNA remained constant in all of the samples (Fig. 1A). The induction of IL-8 was mediated through TLR2 because it was induced only in TLR2 (Fig. 1A) and not in control vector or TLR1 or TLR4 transfectants (data not shown).
FIG. 1.
Micrococci (Micro) and sPGN induce transcription of IL-8 in HEK293 cells expressing TLR2, and this induction is NF-κB dependent. (A) HEK293/TLR2 cells were stimulated with micrococci at 40 μg/ml or sPGN at 10 μg/ml, total RNA was isolated, IL-8 and GAPDH cDNAs were synthesized and amplified using primers specific for IL-8 and GAPDH, and the amplified products were separated on a 3% agarose gel. The results shown are from one of two similar experiments. (B) 293 cells were cotransfected with the following plasmids: TLR2, TLR1, or a control vector at 0.25 μg/ml, CD14 at 0.025 μg/ml, and IL-8 reporter pIL8(wt)CAT at 0.125 μg/ml. At 24 h after transfection, cells were stimulated with micrococci at 40 μg/ml, sPGN at 10 μg/ml, or TNF-α at 100 ng/ml for 16 h and cell lysates were assayed for CAT activity. (C) 293 cells were cotransfected with plasmids TLR2, CD14, and pIL8(wt)CAT (as described for panel B) or with an IL-8 reporter plasmid with a nonfunctional NF-κB site, pIL8(−κB)CAT, at 0.125 μg/ml. Cells were stimulated and assayed as described for panel B. The results are means of duplicate samples from one of three similar experiments. CV, control vector.
We confirmed this induction of IL-8 transcription using an IL-8 promoter–CAT construct, pIL8(wt)CAT, which has bp −420 to +101 of the IL-8 promoter fused to the gene for CAT (17). Micrococci and sPGN strongly induced IL-8 promoter activity, as shown by an increase in CAT activity (Fig. 1B). The average fold induction for micrococci and sPGN was 18.6 and 7.8, respectively. Micrococcus- and sPGN-induced transcription of IL-8 was mediated through TLR2, because this induction was observed in HEK293 cells expressing TLR2 and not in cells expressing the control vector or TLR1 (Fig. 1B) or TLR4 (data not shown).
The IL-8 promoter has a binding site for NF-κB, one of the main transcription factors involved in the regulation of genes during inflammatory and immune responses. To determine whether NF-κB is required for micrococcus- and sPGN-induced IL-8 transcription, we next tested plasmid pIL8 (−κB)CAT (17), which contains a nonfunctional κB site in the IL-8 promoter. Induction of IL-8 transcription by micrococci and sPGN was completely inhibited by a nonfunctional κB site within the IL-8 promoter (Fig. 1C), which indicates that NF-κB is required for induction of the gene for IL-8 by micrococci and sPGN.
Micrococci and sPGN induce binding to an NF-κB site and transactivation of an NF-κB-regulated gene in HEK293 cells expressing TLR2 but not in those expressing TLR1.
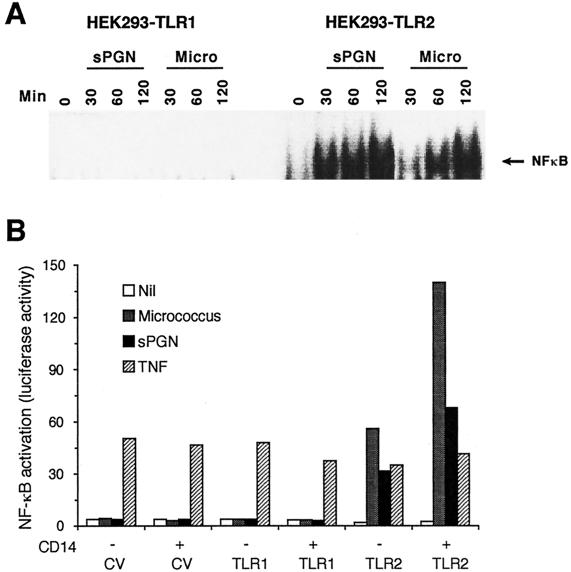
To begin to identify signal transduction pathways that regulate IL-8 induction by gram-positive bacteria and PGN, we first determined the signal transduction pathway involved in the activation of NF-κB. We have recently shown that sPGN induces TLR2-dependent activation of NF-κB (27, 36). In the current experiments, we confirmed these results and tested whether micrococci also induce TLR2-dependent NF-κB activity by two different assays: (i) electrophoretic mobility shift assay of HEK293 cells stably transfected with TLR1, TLR2, or TLR4 and (ii) NF-κB transcriptional activity in HEK293 cells transiently transfected with an NF-κB luciferase plasmid (ELAM-1 luciferase) and with TLR1, TLR2, or TLR4. Cells expressing TLR2, but not those expressing TLR1 (or TLR4 [not shown]), became highly responsive to both micrococci and PGN. Nuclear extracts from activated cells contained proteins that bind to the NF-κB site, as shown by a shift in the band (Fig. 2A). Furthermore, micrococci and sPGN induced NF-κB-dependent luciferase activity in HEK293-TLR2 cells (Fig. 2B), and this activity was enhanced by CD14 (Fig. 2B), although CD14 is not necessary for the responsiveness to micrococci or sPGN (Fig. 2B, and reference 27). By contrast, cells transfected with a control vector or TLR1 (or TLR4 [not shown]) were unresponsive to micrococci or sPGN, regardless of the presence of CD14. These data indicate that TLR2 is a cell-activating receptor for both micrococci and sPGN. As expected, expression of TLRs did not influence TNF-induced activation of NF-κB (Fig. 2B).
FIG. 2.
Micrococci (Micro) and sPGN activate NF-κB in HEK293 cells expressing TLR2 but not in cells expressing TLR1. (A) HEK293 cells expressing TLR1 or TLR2 were stimulated as described in the legend to Fig. 1A for the indicated times, and NF-κB was detected in nuclear extracts by electrophoretic mobility shift assay. The results shown are from one of two similar experiments. (B) HEK293 cells were transfected with the following plasmids: TLR1, TLR2, or a control vector (CV) and CD14 as described in the legend to Fig. 1B and the NF-κB reporter plasmid ELAM-1 luciferase at 0.125 μg/ml. Cells were stimulated as described in the legend Fig. 1B for 6 h, and cell lysates were assayed for luciferase activity. The results are means of duplicate samples from one of two similar experiments.
Micrococcus- and sPGN-induced TLR2-dependent NF-κB activation is mediated by MyD88, IRAK, TRAF6, NIK, and IKK.
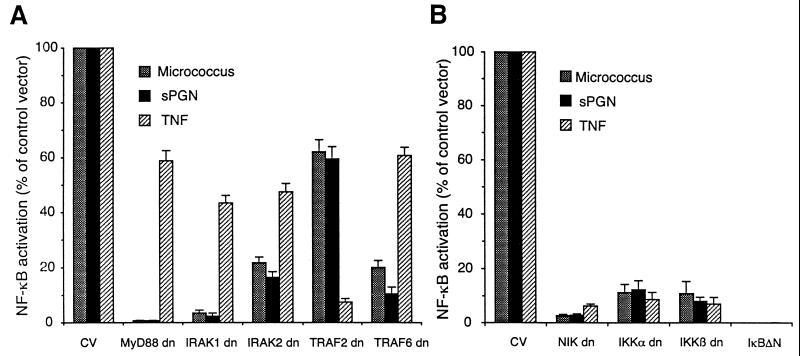
Because the C-terminal regions of TLR2 and IL-1R share significant sequence homology and because the LPS-induced TLR2-mediated signal transduction pathway resembles the IL-1-induced signal transduction pathway (16, 35, 37), we tested whether this signal transduction pathway is also involved in TLR2-induced NF-κB activation by gram-positive bacteria and sPGN. We cotransfected cells with TLR2, CD14, and NF-κB reporter plasmids and dominant negative mutant forms of various components of the above signal transduction pathways. Dominant negative MyD88 and dominant negative IRAK1 completely inhibited micrococcus- and sPGN-induced NF-κB activity in 293 cells expressing TLR2 (Fig. 3A). There was also significant inhibition of micrococcus- and PGN-induced NF-κB activity by dominant negative IRAK2 and TRAF6 (Fig. 3A). As expected, TRAF2 (a component of the TNF-R signal transduction pathway), but not TRAF6, almost completely inhibited TNF-α-induced NF-κB activation (Fig. 3A). Inhibition by these plasmids was dose dependent (data not shown). These results indicate participation of MyD88, IRAK, and TRAF6 in the signal transduction pathway activated by PGN and micrococci.
FIG. 3.
Micrococcus- and sPGN-induced NF-κB activation in HEK293 cells expressing TLR2 is inhibited by dominant negative (dn) MyD88, IRAK, TRAF6, NIK, and IKK. HEK293 cells were cotransfected with plasmids expressing TLR2, CD14, and ELAM-1 luciferase as described in the legend to Fig. 2B and with dominant negative mutant forms of the following signal transduction molecules: MyD88, IRAK1, or IRAK2 at 1.25 μg/ml; TRAF2 or TRAF6 at 0.625 μg/ml; or the appropriate control vector(s); (CV) (A) and NIK, IKKα, IKKβ, IκBΔN, or the appropriate control vector(s) at 1.25 μg/ml (B). Cells were stimulated, and cell lysates were assayed for luciferase activity (LA). The following equation was used: percentage of control vector = (mean LA stimulated with dominant negative − mean LA unstimulated with dominant negative) × 100/(mean LA stimulated with empty vector − mean LA unstimulated with empty vector). The results shown are means ± standard errors from three experiments.
In the IL-1 signaling pathway, activation of TRAF6 results in phosphorylation and activation of NIK, which stimulates IKK activity. Activated IKK phosphorylates IκB, which results in degradation of this inhibitory protein and subsequent release and activation of NF-κB (33). We tested the role of the kinases NIK, IKKα, and IKKβ in micrococcus- and sPGN-induced NF-κB activation in cells expressing TLR2. Dominant negative NIK, IKKα, and IKKβ completely inhibited NF-κB activation by all three stimulants (Fig. 3B), indicating that these kinases are required for NF-κB activation by micrococci, sPGN, and TNF-α. A constitutive repressor of IκB (the inhibitory protein for NF-κB), IκBΔN, also completely inhibited NF-κB activation by micrococci, sPGN, and TNF-α (Fig. 3B), further confirming that degradation of IκB is necessary for activation of NF-κB.
Micrococcus- and sPGN-induced TLR2-dependent IL-8 expression is mediated by MyD88, IRAK, TRAF6, TRAF2, NIK, and IKK.
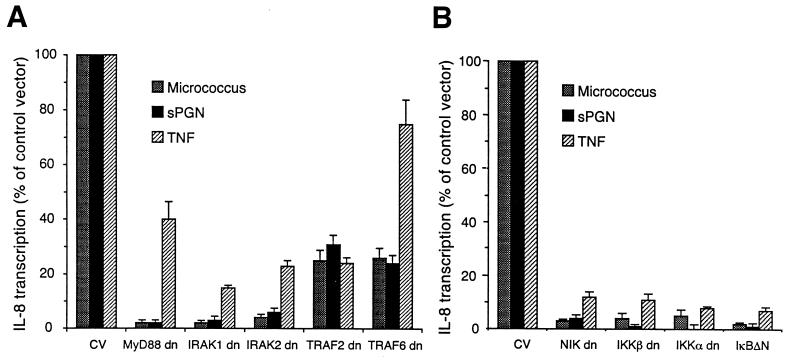
Because activation of NF-κB in cells stimulated with micrococci and sPGN was mediated through the signal transduction molecules MyD88, IRAK, and TRAF6, we next tested if induction of IL-8 was also mediated through these signal transduction molecules. Dominant negative forms of MyD88 and IRAK completely inhibited and dominant negative forms of TRAF6 and TRAF2 partially inhibited induction of IL-8 transcription by micrococci and sPGN (Fig. 4A). TNF-α-induced expression of IL-8 was inhibited by dominant negative IRAK1, IRAK2, and TRAF2, partially inhibited by dominant negative MyD88, and not inhibited by dominant negative TRAF6.
FIG. 4.
Micrococcus- and sPGN-induced transcription of the gene for IL-8 in HEK293 cells expressing TLR2 is inhibited by dominant negative (dn) MyD88, IRAK, TRAF2, TRAF6, NIK, and IKK. HEK293 cells were cotransfected with plasmids expressing TLR2, CD14, and pIL8(wt)CAT and with dominant negative mutant forms of the following signal transduction molecules: MyD88, IRAK1, IRAK2, TRAF2, TRAF6, or the appropriate control vector (CV) (A) and NIK, IKKα, IKKβ, IκBΔN, or the appropriate control vector (B). All plasmids were used at the same concentrations as in previous experiments. Cells were stimulated as described in the legend to Fig. 1 for 16 h, and cell lysates were assayed for CAT activity. The results were calculated as described in the legend to Fig. 3 and are means ± standard errors from three experiments.
We next determined the requirement for the kinases NIK, IKKα, and IKKβ in micrococcus- and sPGN-induced expression of the gene for IL-8 in 293-TLR2 cells. Dominant negative forms of NIK, IKKα, and IKKβ strongly inhibited transcriptional induction of the gene for IL-8 by all three stimulants (Fig. 4B). These data indicate that NIK, IKKα, and IKKβ are required for TLR2-dependent induction of expression of the gene for IL-8 by micrococci and sPGN.
DISCUSSION
Our results demonstrate that (i) micrococci and sPGN induce transcription of the gene for IL-8 in cells expressing TLR2 but not in cells expressing TLR1; (ii) micrococci and sPGN induce activation of NF-κB in cells expressing TLR2, and this activation requires the signal transduction molecules MyD88, IRAK1, NIK, IKKα, and IKKβ and, to a lesser extent, IRAK2 and TRAF6; and (iii) micrococcus- and sPGN-induced transcription of the gene for IL-8 requires MyD88, IRAK1, IRAK2, NIK, IKKα, IKKβ, and NF-κB and, to a lesser extent, TRAF6 and TRAF2. These data are the first to identify signal transduction molecules required for IL-8 induction in cells stimulated with bacteria and bacterial cell wall products.
These data also confirm our previous results showing that gram-positive bacteria and bacterial cell wall component PGN activate cells through TLR2, but not TLR1 or TLR4 (8, 27, 36), and that this TLR2-mediated cell activation is enhanced by CD14 (27). PGN binds CD14 (5), and this binding may be the first step in PGN-induced cell activation. Although we have previously shown indirect activation of endothelial and epithelial cells by PGN-induced TNF-α and IL-1 secreted from monocytes (15), in our current experiments, activation of 293 cells by PGN is a direct activation mediated through TLR2 and CD14 and not an indirect effect because 293 cells do not produce TNF-α and IL-1 (D. Gupta and R. Dziarski, unpublished data).
Of the six different TLRs that have been identified (published), only two have a known function, TLR2 and TLR4. TLR2 is a cell-activating receptor for gram-positive bacteria (8, 27, 29, 36), mycobacteria (2, 18), spirochetes (13), and mycoplasmas (18) and for the cell wall components PGN (8, 29, 27, 36), lipoteichoic acid (8, 27, 29), lipopeptides, lipoproteins (2, 13), and ara-lipoarabinomannan (19). Moreover, an accessory molecule, MD-2 (28), enables TLR2 to respond to various nonactivating LPS structures (8). TLR2 is recruited to macrophage phagosomes containing yeast, and dominant negative TLR2 abolishes TNF-α production in response to yeast and to gram-positive bacteria but not in response to gram-negative bacteria (31). Thus, TLR2 appears to be a true pattern recognition receptor that recognizes a large variety of microbes and microbial products and mediates cell activation and host inflammatory responses. However, a structural feature common to these putative ligands that is needed for recognition by TLR2 has not been identified.
TLR4 is implicated in host immune responses to gram-negative bacteria and to their LPS cell wall component (22, 23, 29) but not to gram-positive bacteria or PGN (8, 29, 30). The Lps gene in C3H/HeJ and C57BL/10ScCr mice, which is responsible for their defective responsiveness to LPS, was identified as Tlr4 (22, 23). Moreover, TLR4-mediated cell activation by LPS requires coexpression of the accessory molecule MD-2 (8, 28).
Our data are the first to demonstrate that the signal transduction molecules MyD88, IRAK, NIK, and IKK are required for TLR2-mediated NF-κB activation in cells stimulated with micrococci and bacterial PGN (Fig. 3). These data demonstrate that gram-positive bacteria and their cell wall components induce TLR2-mediated signal transduction pathways that are similar to the TLR-dependent LPS-induced signal transduction pathways and involve IRAK, MyD88, TRAF6, IKKα, IKKβ, NIK, and NF-κB (16, 35, 37) and confirm the role of MyD88 as an essential component in this pathway (30). This pathway is also similar to the IL-1-induced signal transduction pathway (4, 20), which is consistent with the homology of the cytoplasmic domains of TLR2 and IL-1R.
Our data also demonstrate TLR2-mediated transcriptional induction of IL-8 by micrococci and PGN and the requirement for NF-κB in induction of IL-8 by gram-positive bacterial stimulants. Although NF-κB is required for the induction of IL-8 transcription, it may not be sufficient, and other signal transduction pathways may also be required or may modulate IL-8 transcription. This study was done on a human embryonic kidney cell line, and this cell line, as many other cells in the body, such as endothelial cells, epithelial cells, and fibroblasts, can produce IL-8, in addition to the well-known production of IL-8 by bacterially activated monocytes (32). However, the signal transduction pathways and transcription factors needed for activation of transcription of the gene for IL-8 are likely to be the same in all of these cells because the IL-8 promoter is the same in all of the cells. Therefore, our results should be applicable to all cells that are able to produce IL-8, and their responsiveness to bacterial products would depend on the expression of appropriate receptors, such as CD14 and TLR2.
The signal transduction molecules MyD88, IRAK, NIK, and IKK are required for TLR2-dependent induction of IL-8 expression in cells stimulated with micrococci and PGN. Both dominant negative TRAF6 and TRAF2 partially inhibit IL-8 induction in response to these bacterial products. Therefore, these results suggest that alternative pathways involving TRAF2 or TRAF6 may play an equal role in IL-8 induction, but not in NF-κB induction, in cells stimulated with bacteria and bacterial cell wall components (Fig. 5). However, there may also be cross talk between TNF-R- and TLR2–IL-1R-induced pathways because of the partial inhibition of TNF-induced IL-8 activation by dominant negative MyD88 and IRAK.
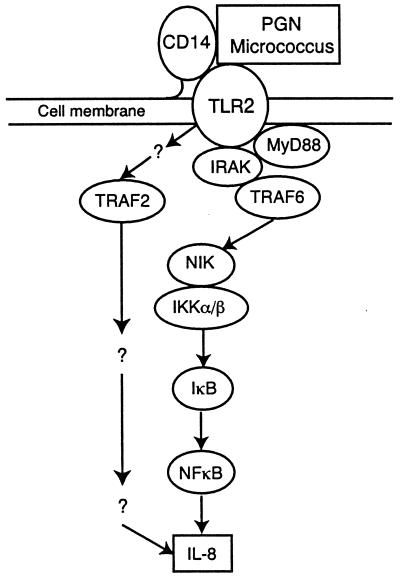
FIG. 5.
Proposed signal transduction pathway activated by micrococci and sPGN.
In summary, our results demonstrate that gram-positive bacteria and PGN induce IL-8 transcription through the TLR2→MyD88→IRAK→TRAF6→NIK→IKK→NF-κB (i.e., IL-1R-like) signal transduction pathway, although additional signal transduction molecules may participate in this pathway, and other signal transduction pathways may also be involved in induction of IL-8 transcription.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant AI28797 (to R.D).
We are grateful to David Goeddel for providing the TRAF6(289-522), TRAF2(87-501), NIK(KK429, 430AA), IKKα(S176A), and IKKβ(S177A) plasmids; Charles Kunsch for the IL-8 reporter plasmids; and D. W. Ballard for the IκBΔN plasmid.
REFERENCES
- 1.Bone R C. Gram-positive organisms and sepsis. Arch Intern Med. 1994;154:26–34. [PubMed] [Google Scholar]
- 2.Brightbill H D, Libraty D H, Krutzik S R, Yang R B, Belisle J T, Bleharski J R, Maitland M, Norgard M V, Plevy S E, Smale S T, Brennan P J, Bloom B R, Godowski P J, Modlin R L. Host defense mechanisms triggered by microbial lipoproteins through Toll-like receptors. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 3.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Coupling of a signal response domain in IκBα to multiple pathways for NF-κB activation. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel D V. TRAF6 is a signal transducer for interleukin-1. Science. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 5.Dziarski R, Tapping R I, Tobias P S. Binding of bacterial peptidoglycan to CD14. J Biol Chem. 1998;273:8680–8690. doi: 10.1074/jbc.273.15.8680. [DOI] [PubMed] [Google Scholar]
- 6.Dziarski R, Ulmer A, Gupta D. Interactions of bacterial lipopolysaccharide and peptidoglycan with mammalian CD14. In: Doyle R J, editor. Glycomicrobiology. New York, N.Y: Kluwer Academic/Plenum Publishers; 2000. pp. 145–186. [Google Scholar]
- 7.Dziarski R, Ulmer A, Gupta D. Interactions of CD14 with components of Gram-positive bacteria. Chem Immunol. 2000;74:83–107. doi: 10.1159/000058761. [DOI] [PubMed] [Google Scholar]
- 8.Dziarski R, Wang Q, Kirschning C J, Miyake K, Gupta D. MD-2 enables Toll-like receptor-2 (TLR2)-mediated responses to LPS and enhances TLR2-mediated responses to Gram-positive and Gram-negative bacteria and their cell wall components. J Immunol. 2001;166:1938–1944. doi: 10.4049/jimmunol.166.3.1938. [DOI] [PubMed] [Google Scholar]
- 9.Feldman M, Brennan F M, Maini R N. Role of cytokines in rheumatoid arthritis. Annu Rev Immunol. 1996;14:397–440. doi: 10.1146/annurev.immunol.14.1.397. [DOI] [PubMed] [Google Scholar]
- 10.Fischer C, Page S, Weber M, Eisele T, Neumeier D, Brand K. Differential activation of lipolpolysaccharide and tumor necrosis factor on monocytic IκB kinase signalsome activation and IκB proteolysis. J Biol Chem. 1999;274:24625–24632. doi: 10.1074/jbc.274.35.24625. [DOI] [PubMed] [Google Scholar]
- 11.Gupta D, Kirkland T N, Viriyakosol S, Dziarski R. CD14 is a cell-activating receptor for bacterial peptidoglycan. J Biol Chem. 1996;271:23310–23316. doi: 10.1074/jbc.271.38.23310. [DOI] [PubMed] [Google Scholar]
- 12.Gupta D, Wang Q, Vinson C, Dziarski R. Bacterial peptidoglycan induces CD14-dependent activation of transcription factors CREB/ATF and AP-1. J Biol Chem. 1999;274:14012–14020. doi: 10.1074/jbc.274.20.14012. [DOI] [PubMed] [Google Scholar]
- 13.Hirschfeld M, Kirschning C J, Schwander R, Wesche H, Weis J H, Wooten R M, Weis J J. Inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by Toll-like receptor 2. J Immunol. 1999;163:2382–2386. [PubMed] [Google Scholar]
- 14.Izutani R, Ohyanagi H, MacDermott R. Quantitative PCR for detection of femtogram quantities of interleukin-8 mRNA expression. Microbiol Immunol. 1994;38:233–237. doi: 10.1111/j.1348-0421.1994.tb01770.x. [DOI] [PubMed] [Google Scholar]
- 15.Jin Y, Gupta D, Dziarski R. Endothelial and epithelial cells do not respond to complexes of peptidoglycan with soluble CD14 but are activated indirectly by peptidoglycan-induced tumor necrosis factor-α and interleukin-1 from monocytes. J Infect Dis. 1998;177:1629–1638. doi: 10.1086/515318. [DOI] [PubMed] [Google Scholar]
- 16.Kirschning C J, Wesche H, Ayres T M, Rothe M. Human Toll-like receptor 2 confers responsiveness to bacterial lipopolysaccharide. J Exp Med. 1998;188:2091–2097. doi: 10.1084/jem.188.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunsch C, Lang R K, Rosen C A, Shannon M. Synergistic transcriptional activation of IL-8 by NF-kappa B p65 (RelA) and NF-IL-6. J Immunol. 1994;153:153–164. [PubMed] [Google Scholar]
- 18.Lien E, Sellati T J, Yoshimura A, Flo T H, Rawadi G, Finberg R W, Carroll J D, Espevik T, Ingalls R, Radolf J D, Golenbock D T. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J Biol Chem. 1999;274:33419–33425. doi: 10.1074/jbc.274.47.33419. [DOI] [PubMed] [Google Scholar]
- 19.Means T K, Wang S, Lien E, Yoshimura A, Golenbock D T, Fenton M. Human Toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. J Immunol. 1999;163:3920–3927. [PubMed] [Google Scholar]
- 20.Muzio M, Ni J, Feng P, Dixit V. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278:1612–1615. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- 21.Parillo J E. Pathogenic mechanisms of septic shock. N Engl J Med. 1993;328:1471–1477. doi: 10.1056/NEJM199305203282008. [DOI] [PubMed] [Google Scholar]
- 22.Poltorak A, He X, Smirnova I, Liu M-Y, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Riccardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 23.Qureshi S T, Lariviere L, Leveque G, Clermont S, Moore K J, Gros P, Malo D. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4) J Exp Med. 1999;189:615–625. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reigner C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 25.Rosenthal R S, Dziarski R. Isolation of peptidoglycan and soluble peptidoglycan fragments. Methods Enzymol. 1994;235:253–285. doi: 10.1016/0076-6879(94)35146-5. [DOI] [PubMed] [Google Scholar]
- 26.Rothe M, Wong S C, Henzel W J, Goeddel D V. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 27.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning C J. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by Toll-like receptor 2. J Biol Chem. 1999;274:17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 28.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of Gram-negative and Gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi O, Takeda K, Hoshino K, Adachi O, Ogawa T, Akira S. Cellular responses to bacterial cell wall components are mediated through MyD88-dependent signaling cascades. Int Immunol. 2000;12:113–117. doi: 10.1093/intimm/12.1.113. [DOI] [PubMed] [Google Scholar]
- 31.Underhill D M, Ozinsky A, Hajjar A M, Stevens A, Wilson C B, Bassetti M, Aderem A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;40:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z-M, Liu C, Dziarski R. Chemokines are the main pro-inflammatory mediators in human monocytes activated by Staphylococcus aureus, peptidoglycan, and endotoxin. J Biol Chem. 2000;275:20260–20267. doi: 10.1074/jbc.M909168199. [DOI] [PubMed] [Google Scholar]
- 33.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 34.Yang R-B, Mark M R, Gray A, Huang A, Xie M H, Zhang M, Goddard A, Wood W I, Gurney A L, Godowski P J. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signaling. Nature. 1998;395:284–288. doi: 10.1038/26239. [DOI] [PubMed] [Google Scholar]
- 35.Yang R-B, Mark M R, Gurney A L, Godowski P J. Signaling events induced by lipopolysaccharide-activated Toll-like receptor 2. J Immunol. 1999;163:639–643. [PubMed] [Google Scholar]
- 36.Yoshimura A, Lien E, Ingalls R R, Tuomanen E, Dziarski R, Golenbock D. Recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- 37.Zhang F X, Kirschning C J, Mancinelli R, Xu X-P, Jin Y, Faure E, Mantovani A, Rothe M, Muzio M, Arditi M. Bacterial lipopolysaccharide activates nuclear factor-κB through interleukin-1 signaling mediators in cultured human dermal endothelial cells and mononuclear phagocytes. J Biol Chem. 1999;274:7611–7614. doi: 10.1074/jbc.274.12.7611. [DOI] [PubMed] [Google Scholar]